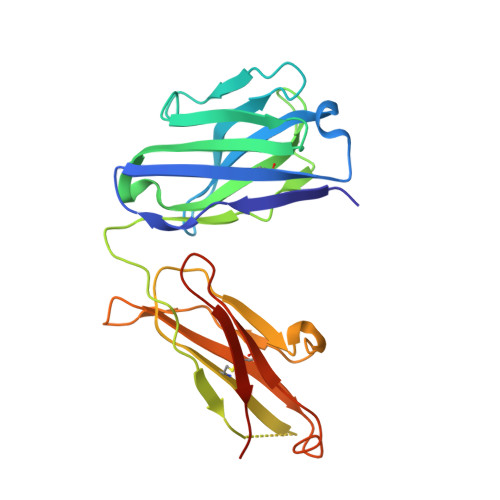
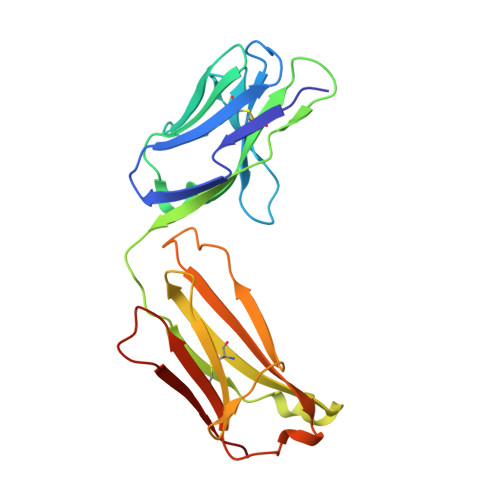
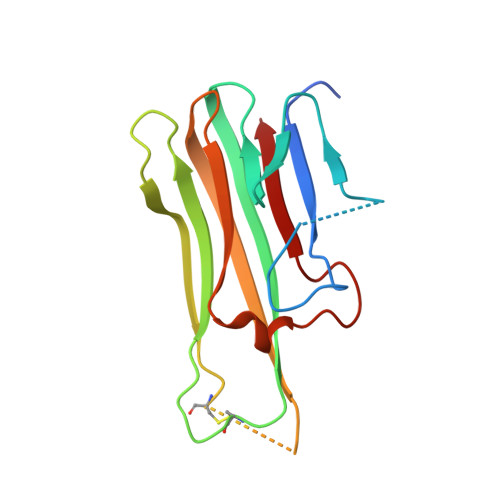
Molecular Basis for the Neutralization of Tumor Necrosis Factor alpha by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases
Lee, J.U., Shin, W., Son, J.Y., Yoo, K.Y., Heo, Y.S.(2017) Int J Mol Sci 18
- PubMed: 28124979
- DOI: https://doi.org/10.3390/ijms18010228
- Primary Citation of Related Structures:
5WUV, 5WUX - PubMed Abstract:
Monoclonal antibodies against TNFα, including infliximab, adalimumab, golimumab, and certolizumab pegol, are widely used for the treatment of the inflammatory diseases such as rheumatoid arthritis and inflammatory bowel disease. Recently, the crystal structures of TNFα, in complex with the Fab fragments of infliximab and adalimumab, have revealed the molecular mechanisms of these antibody drugs. Here, we report the crystal structure of TNFα in complex with the Fab fragment of certolizumab pegol to clarify the precise antigen-antibody interactions and the structural basis for the neutralization of TNFα by this therapeutic antibody. The structural analysis and the mutagenesis study revealed that the epitope is limited to a single protomer of the TNFα trimer. Additionally, the DE loop and the GH loop of TNFα play critical roles in the interaction with certolizumab, suggesting that this drug exerts its effects by partially occupying the receptor binding site of TNFα. In addition, a conformational change of the DE loop was induced by certolizumab binding, thereby interrupting the TNFα-receptor interaction. A comprehensive comparison of the interactions of TNFα blockers with TNFα revealed the epitope diversity on the surface of TNFα, providing a better understanding of the molecular mechanism of TNFα blockers. The accumulation of these structural studies can provide a basis for the improvement of therapeutic antibodies against TNFα.
- Department of Chemistry, Konkuk University, 120 Neungdong-ro, Gwangjin-gu, Seoul 05029, Korea. jaspersky@naver.com.
Organizational Affiliation: