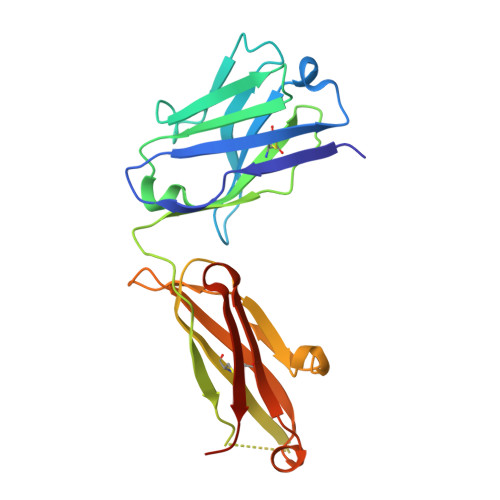
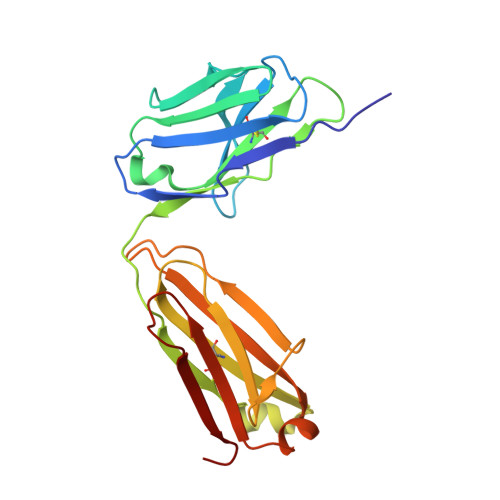
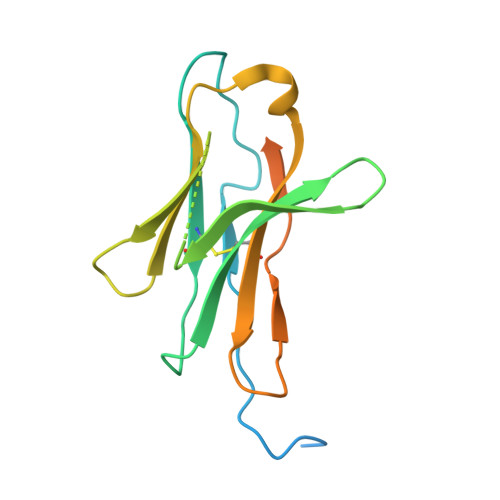
An unexpected N-terminal loop in PD-1 dominates binding by nivolumab.
Tan, S., Zhang, H., Chai, Y., Song, H., Tong, Z., Wang, Q., Qi, J., Wong, G., Zhu, X., Liu, W.J., Gao, S., Wang, Z., Shi, Y., Yang, F., Gao, G.F., Yan, J.(2017) Nat Commun 8: 14369-14369
- PubMed: 28165004
- DOI: https://doi.org/10.1038/ncomms14369
- Primary Citation of Related Structures:
5WT9 - PubMed Abstract:
Cancer immunotherapy by targeting of immune checkpoint molecules has been a research 'hot-spot' in recent years. Nivolumab, a human monoclonal antibody targeting PD-1, has been widely used clinically since 2014. However, the binding mechanism of nivolumab to PD-1 has not yet been shown, despite a recent report describing the complex structure of pembrolizumab/PD-1. It has previously been speculated that PD-1 glycosylation is involved in nivolumab recognition. Here we report the complex structure of nivolumab with PD-1 and evaluate the effects of PD-1 N-glycosylation on the interactions with nivolumab. Structural and functional analyses unexpectedly reveal an N-terminal loop outside the IgV domain of PD-1. This loop is not involved in recognition of PD-L1 but dominates binding to nivolumab, whereas N-glycosylation is not involved in binding at all. Nivolumab binds to a completely different area than pembrolizumab. These results provide the basis for the design of future inhibitory molecules targeting PD-1.
- CAS Key Laboratory of Microbial Physiological and Metabolic engineering, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: