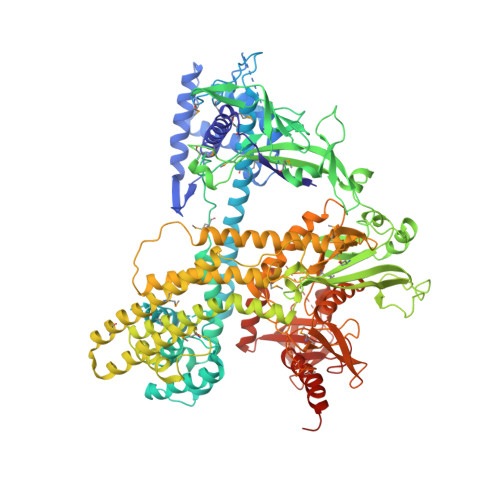
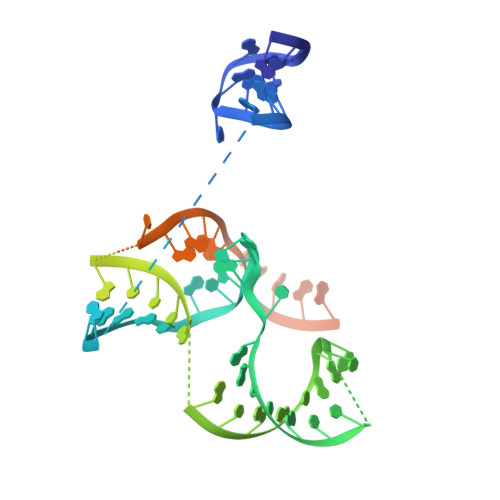
C2c1-sgRNA Complex Structure Reveals RNA-Guided DNA Cleavage Mechanism
Liu, L., Chen, P., Wang, M., Li, X., Wang, J., Yin, M., Wang, Y.(2017) Mol Cell 65: 310-322
- PubMed: 27989439
- DOI: https://doi.org/10.1016/j.molcel.2016.11.040
- Primary Citation of Related Structures:
5WQE - PubMed Abstract:
C2c1 is a type V-B CRISPR-Cas system dual-RNA-guided DNA endonuclease. Here, we report the crystal structure of Alicyclobacillus acidoterrestris C2c1 in complex with a chimeric single-molecule guide RNA (sgRNA). AacC2c1 exhibits a bi-lobed architecture consisting of a REC and NUC lobe. The sgRNA scaffold forms a tetra-helical structure, distinct from previous predictions. The crRNA is located in the central channel of C2c1, and the tracrRNA resides in an external surface groove. Although AacC2c1 lacks a PAM-interacting domain, our analysis revealed that the PAM duplex has a similar binding position found in Cpf1. Importantly, C2c1-sgRNA system is highly sensitive to single-nucleotide mismatches between guide RNA and target DNA. The resulting reduction in off-target cleavage renders C2c1 a valuable addition to the current arsenal of genome-editing tools. Together, our findings indicate that sgRNA assembly is achieved through a mechanism distinct from that reported previously for Cas9 or Cpf1 endonucleases.
- Key Laboratory of RNA Biology, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: