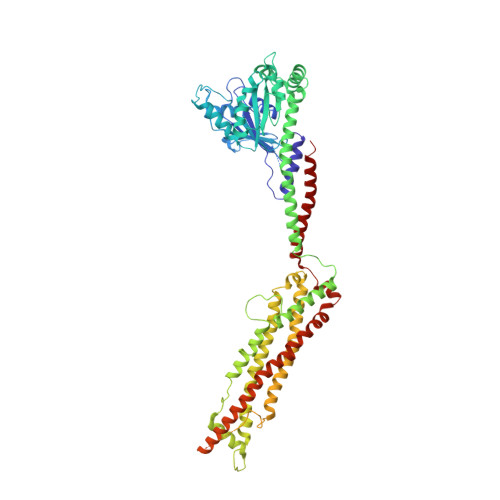
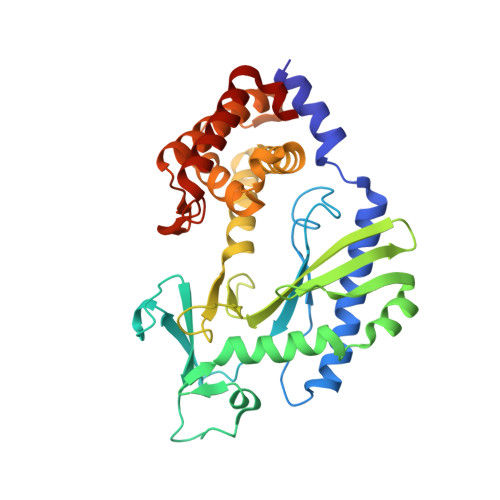
Structural basis of mitochondrial receptor binding and constriction by DRP1.
Kalia, R., Wang, R.Y., Yusuf, A., Thomas, P.V., Agard, D.A., Shaw, J.M., Frost, A.(2018) Nature 558: 401-405
- PubMed: 29899447
- DOI: https://doi.org/10.1038/s41586-018-0211-2
- Primary Citation of Related Structures:
5WP9 - PubMed Abstract:
Mitochondrial inheritance, genome maintenance and metabolic adaptation depend on organelle fission by dynamin-related protein 1 (DRP1) and its mitochondrial receptors. DRP1 receptors include the paralogues mitochondrial dynamics proteins of 49 and 51 kDa (MID49 and MID51) and mitochondrial fission factor (MFF); however, the mechanisms by which these proteins recruit and regulate DRP1 are unknown. Here we present a cryo-electron microscopy structure of full-length human DRP1 co-assembled with MID49 and an analysis of structure- and disease-based mutations. We report that GTP induces a marked elongation and rotation of the GTPase domain, bundle-signalling element and connecting hinge loops of DRP1. In this conformation, a network of multivalent interactions promotes the polymerization of a linear DRP1 filament with MID49 or MID51. After co-assembly, GTP hydrolysis and exchange lead to MID receptor dissociation, filament shortening and curling of DRP1 oligomers into constricted and closed rings. Together, these views of full-length, receptor- and nucleotide-bound conformations reveal how DRP1 performs mechanical work through nucleotide-driven allostery.
- Department of Biochemistry and Biophysics, University of California, San Francisco, San Francisco, CA, USA.
Organizational Affiliation: