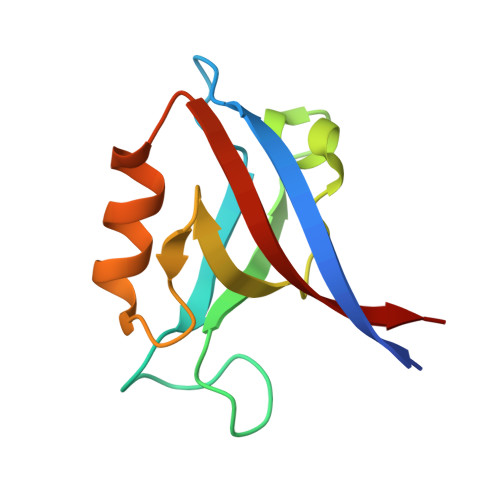
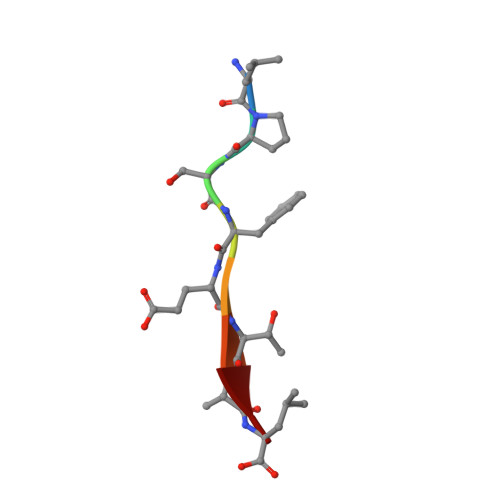
Drosophila melanogasterGuk-holder interacts with the Scribbled PDZ1 domain and regulates epithelial development with Scribbled and Discs Large.
Caria, S., Magtoto, C.M., Samiei, T., Portela, M., Lim, K.Y.B., How, J.Y., Stewart, B.Z., Humbert, P.O., Richardson, H.E., Kvansakul, M.(2018) J Biological Chem 293: 4519-4531
- PubMed: 29378849
- DOI: https://doi.org/10.1074/jbc.M117.817528
- Primary Citation of Related Structures:
5WOU - PubMed Abstract:
Epithelial cell polarity is controlled by components of the Scribble polarity module, and its regulation is critical for tissue architecture and cell proliferation and migration. In Drosophila melanogaster , the adaptor protein Guk-holder (Gukh) binds to the Scribbled (Scrib) and Discs Large (Dlg) components of the Scribble polarity module and plays an important role in the formation of neuromuscular junctions. However, Gukh's role in epithelial tissue formation and the molecular basis for the Scrib-Gukh interaction remain to be defined. We now show using isothermal titration calorimetry that the Scrib PDZ1 domain is the major site for an interaction with Gukh. Furthermore, we defined the structural basis of this interaction by determining the crystal structure of the Scrib PDZ1-Gukh complex. The C-terminal PDZ-binding motif of Gukh is located in the canonical ligand-binding groove of Scrib PDZ1 and utilizes an unusually extensive network of hydrogen bonds and ionic interactions to enable binding to PDZ1 with high affinity. We next examined the role of Gukh along with those of Scrib and Dlg in Drosophila epithelial tissues and found that Gukh is expressed in larval-wing and eye-epithelial tissues and co-localizes with Scrib and Dlg at the apical cell cortex. Importantly, we show that Gukh functions with Scrib and Dlg in the development of Drosophila epithelial tissues, with depletion of Gukh enhancing the eye- and wing-tissue defects caused by Scrib or Dlg depletion. Overall, our findings reveal that Scrib's PDZ1 domain functions in the interaction with Gukh and that the Scrib-Gukh interaction has a key role in epithelial tissue development in Drosophila .
- From the Department of Biochemistry and Genetics, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, Victoria 3086.
Organizational Affiliation: