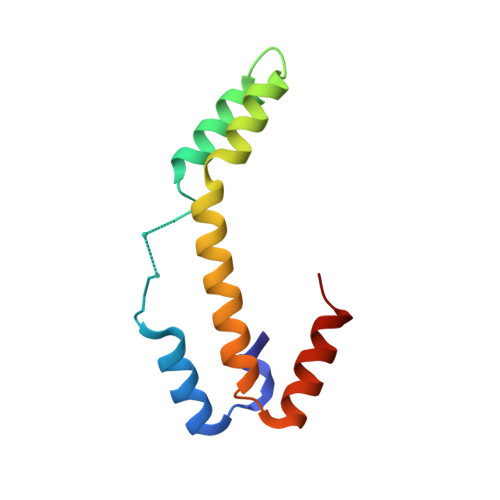
Visualizing chaperone-assisted protein folding.
Horowitz, S., Salmon, L., Koldewey, P., Ahlstrom, L.S., Martin, R., Quan, S., Afonine, P.V., van den Bedem, H., Wang, L., Xu, Q., Trievel, R.C., Brooks, C.L., Bardwell, J.C.(2016) Nat Struct Mol Biol 23: 691-697
- PubMed: 27239796
- DOI: https://doi.org/10.1038/nsmb.3237
- Primary Citation of Related Structures:
5WNW, 5WO1, 5WO2, 5WO3 - PubMed Abstract:
Challenges in determining the structures of heterogeneous and dynamic protein complexes have greatly hampered past efforts to obtain a mechanistic understanding of many important biological processes. One such process is chaperone-assisted protein folding. Obtaining structural ensembles of chaperone-substrate complexes would ultimately reveal how chaperones help proteins fold into their native state. To address this problem, we devised a new structural biology approach based on X-ray crystallography, termed residual electron and anomalous density (READ). READ enabled us to visualize even sparsely populated conformations of the substrate protein immunity protein 7 (Im7) in complex with the Escherichia coli chaperone Spy, and to capture a series of snapshots depicting the various folding states of Im7 bound to Spy. The ensemble shows that Spy-associated Im7 samples conformations ranging from unfolded to partially folded to native-like states and reveals how a substrate can explore its folding landscape while being bound to a chaperone.
- Department of Molecular, Cellular, and Developmental Biology, University of Michigan, Ann Arbor, Michigan, USA.
Organizational Affiliation: