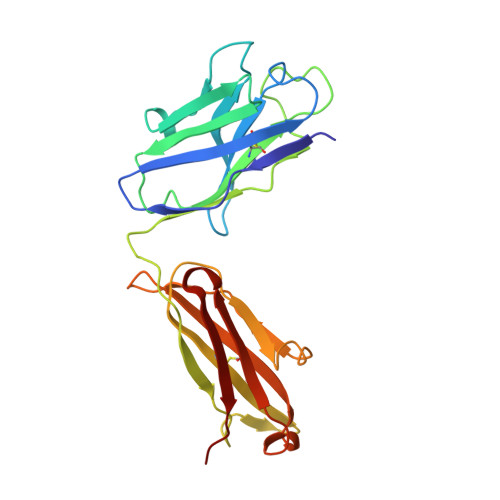
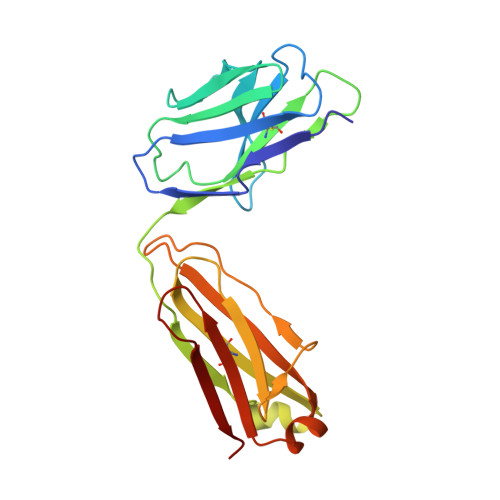
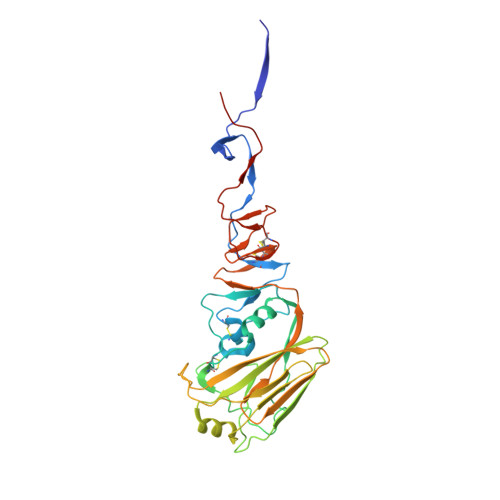
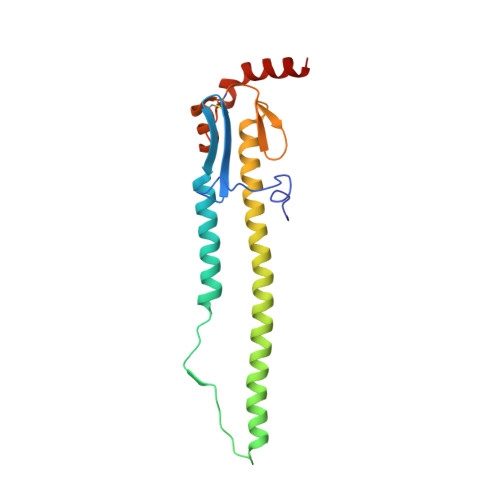
Antibody 27F3 Broadly Targets Influenza A Group 1 and 2 Hemagglutinins through a Further Variation in VH1-69 Antibody Orientation on the HA Stem.
Lang, S., Xie, J., Zhu, X., Wu, N.C., Lerner, R.A., Wilson, I.A.(2017) Cell Rep 20: 2935-2943
- PubMed: 28930686
- DOI: https://doi.org/10.1016/j.celrep.2017.08.084
- Primary Citation of Related Structures:
5WKO - PubMed Abstract:
Antibodies that target both group 1 and group 2 influenza A viruses are valuable for therapeutic and vaccine development, but only a few have been reported to date. Here, we describe a new V H 1-69 antibody 27F3 that broadly recognizes heterosubtypic hemagglutinins (HAs) from both group 1 and group 2 influenza A viruses. Structural characterization of 27F3 Fab with A/California/04/2009 (H1N1) hemagglutinin illustrates that 27F3 shares the key binding features observed in other V H 1-69 antibodies to the HA stem. Compared to other V H 1-69 antibodies, the 27F3 V H domain interacts with the HA stem in a distinct orientation, which alters its epitope and may have influenced its breadth. The diverse rotations of V H 1-69 antibodies on the HA stem epitope highlight the different ways that this antibody family can evolve to broadly neutralize influenza A viruses. These results have important implications for understanding how to elicit broad antibody responses against influenza virus.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA; The Skaggs Institute for Chemical Biology, The Scripps Research Institute, 10550 North Torrey Pines Road, La Jolla, CA 92037, USA.
Organizational Affiliation: