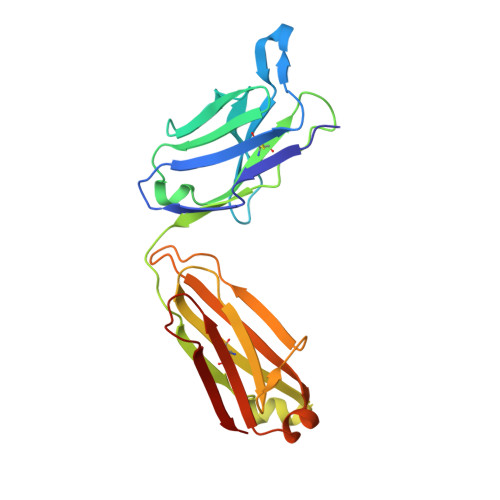
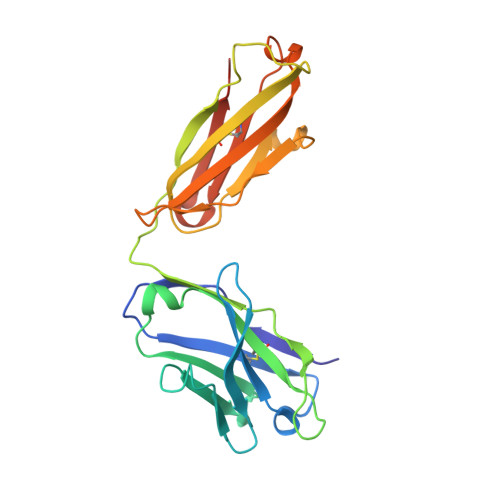
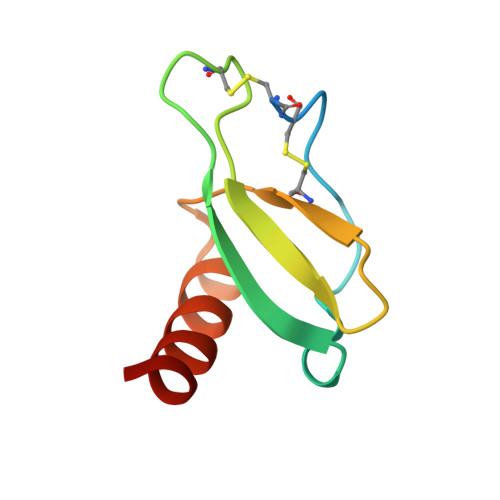
Structural insights into chemokine CCL17 recognition by antibody M116.
Teplyakov, A., Obmolova, G., Gilliland, G.L.(2018) Biochem Biophys Rep 13: 27-31
- PubMed: 29264403
- DOI: https://doi.org/10.1016/j.bbrep.2017.11.005
- Primary Citation of Related Structures:
5WK2, 5WK3 - PubMed Abstract:
The homeostatic chemokine CCL17, also known as thymus and activation regulated chemokine (TARC), has been associated with various diseases such as asthma, idiopathic pulmonary fibrosis, atopic dermatitis and ulcerative colitis. Neutralization of CCL17 by antibody treatment ameliorates the impact of disease by blocking influx of T cells. Monoclonal antibody M116 derived from a combinatorial library shows potency in neutralizing CCL17-induced signaling. To gain insight into the structural determinants of antigen recognition, the crystal structure of M116 Fab was determined in complex with CCL17 and in the unbound form. Comparison of the structures revealed an unusual induced-fit mechanism of antigen recognition that involves cis-trans isomerization in two CDRs. The structure of the CCL17-M116 complex revealed the antibody binding epitope, which does not overlap with the putative receptor epitope, suggesting that the current model of chemokine-receptor interactions, as observed in the CXCR4-vMIP-II system, may not be universal.
- Janssen Research and Development, LLC, Spring House, PA 19477, USA.
Organizational Affiliation: