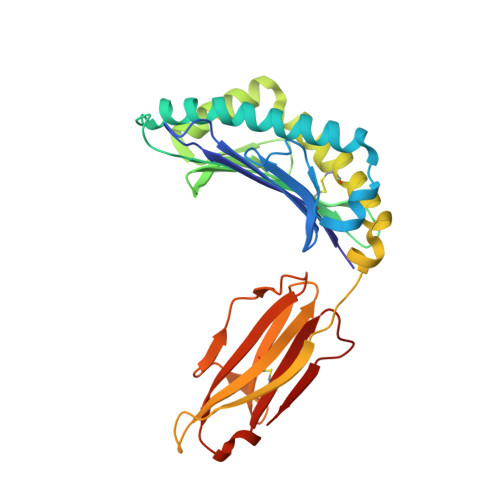
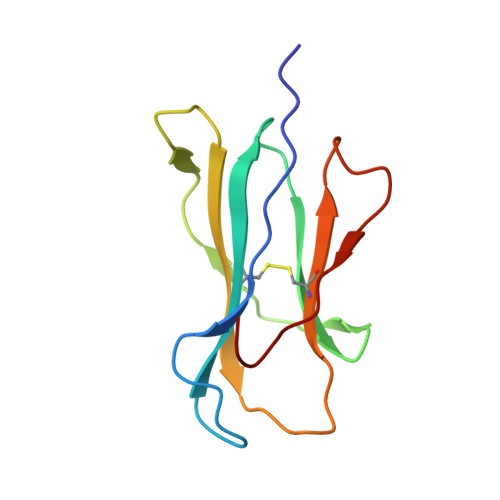
Germline bias dictates cross-serotype reactivity in a common dengue-virus-specific CD8(+) T cell response.
Culshaw, A., Ladell, K., Gras, S., McLaren, J.E., Miners, K.L., Farenc, C., van den Heuvel, H., Gostick, E., Dejnirattisai, W., Wangteeraprasert, A., Duangchinda, T., Chotiyarnwong, P., Limpitikul, W., Vasanawathana, S., Malasit, P., Dong, T., Rossjohn, J., Mongkolsapaya, J., Price, D.A., Screaton, G.R.(2017) Nat Immunol 18: 1228-1237
- PubMed: 28945243
- DOI: https://doi.org/10.1038/ni.3850
- Primary Citation of Related Structures:
5WJL, 5WJN, 5WKF, 5WKH - PubMed Abstract:
Adaptive immune responses protect against infection with dengue virus (DENV), yet cross-reactivity with distinct serotypes can precipitate life-threatening clinical disease. We found that clonotypes expressing the T cell antigen receptor (TCR) β-chain variable region 11 (TRBV11-2) were 'preferentially' activated and mobilized within immunodominant human-leukocyte-antigen-(HLA)-A*11:01-restricted CD8 + T cell populations specific for variants of the nonstructural protein epitope NS3 133 that characterize the serotypes DENV1, DENV3 and DENV4. In contrast, the NS3 133 -DENV2-specific repertoire was largely devoid of such TCRs. Structural analysis of a representative TRBV11-2 + TCR demonstrated that cross-serotype reactivity was governed by unique interplay between the variable antigenic determinant and germline-encoded residues in the second β-chain complementarity-determining region (CDR2β). Extensive mutagenesis studies of three distinct TRBV11-2 + TCRs further confirmed that antigen recognition was dependent on key contacts between the serotype-defined peptide and discrete residues in the CDR2β loop. Collectively, these data reveal an innate-like mode of epitope recognition with potential implications for the outcome of sequential exposure to heterologous DENVs.
- Department of Medicine, Imperial College London, London, UK.
Organizational Affiliation: