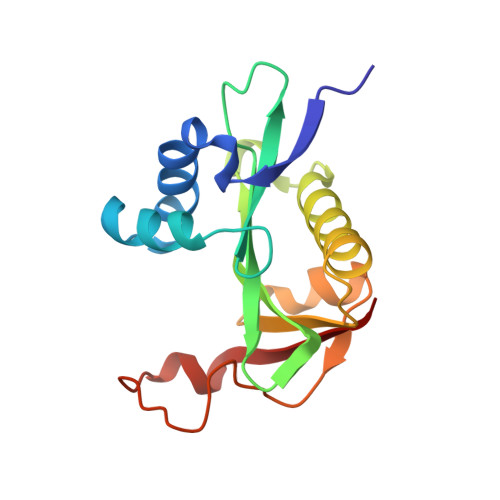
Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80.
Goris, M., Magin, R.S., Foyn, H., Myklebust, L.M., Varland, S., Ree, R., Drazic, A., Bhambra, P., Stove, S.I., Baumann, M., Haug, B.E., Marmorstein, R., Arnesen, T.(2018) Proc Natl Acad Sci U S A 115: 4405-4410
- PubMed: 29581307
- DOI: https://doi.org/10.1073/pnas.1719251115
- Primary Citation of Related Structures:
5WJD, 5WJE - PubMed Abstract:
N-terminal (Nt) acetylation is a major protein modification catalyzed by N-terminal acetyltransferases (NATs). Methionine acidic N termini, including actin, are cotranslationally Nt acetylated by NatB in all eukaryotes, but animal actins containing acidic N termini, are additionally posttranslationally Nt acetylated by NAA80. Actin Nt acetylation was found to regulate cytoskeletal dynamics and motility, thus making NAA80 a potential target for cell migration regulation. In this work, we developed potent and selective bisubstrate inhibitors for NAA80 and determined the crystal structure of NAA80 in complex with such an inhibitor, revealing that NAA80 adopts a fold similar to other NAT enzymes but with a more open substrate binding region. Furthermore, in contrast to most other NATs, the substrate specificity of NAA80 is mainly derived through interactions between the enzyme and the acidic amino acids at positions 2 and 3 of the actin substrate and not residues 1 and 2. A yeast model revealed that ectopic expression of NAA80 in a strain lacking NatB activity partially restored Nt acetylation of NatB substrates, including yeast actin. Thus, NAA80 holds intrinsic capacity to posttranslationally Nt acetylate NatB-type substrates in vivo. In sum, the presence of a dominant cotranslational NatB in all eukaryotes, the specific posttranslational actin methionine removal in animals, and finally, the unique structural features of NAA80 leave only the processed actins as in vivo substrates of NAA80. Together, this study reveals the molecular and cellular basis of NAA80 Nt acetylation and provides a scaffold for development of inhibitors for the regulation of cytoskeletal properties.
- Department of Biological Sciences, University of Bergen, N-5020 Bergen, Norway.
Organizational Affiliation: