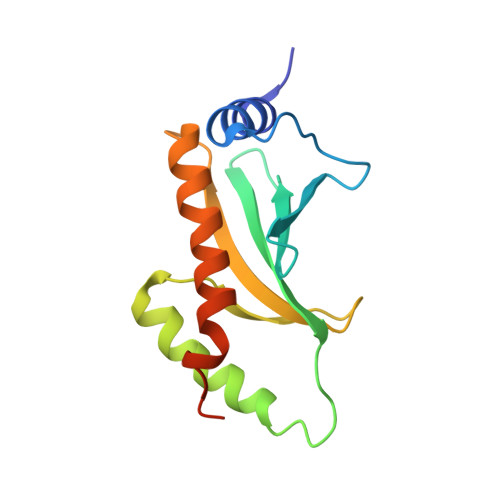
Molecular basis for CesT recognition of type III secretion effectors in enteropathogenic Escherichia coli.
Little, D.J., Coombes, B.K.(2018) PLoS Pathog 14: e1007224-e1007224
- PubMed: 30118511
- DOI: https://doi.org/10.1371/journal.ppat.1007224
- Primary Citation of Related Structures:
5WEZ - PubMed Abstract:
Enteropathogenic Escherichia coli (EPEC) use a needle-like injection apparatus known as the type III secretion system (T3SS) to deliver protein effectors into host cells. Effector translocation is highly stratified in EPEC with the translocated intimin receptor (Tir) being the first effector delivered into the host. CesT is a multi-cargo chaperone that is required for the secretion of Tir and at least 9 other effectors. However, the structural and mechanistic basis for differential effector recognition by CesT remains unclear. Here, we delineated the minimal CesT-binding region on Tir to residues 35-77 and determined the 2.74 Å structure of CesT bound to an N-terminal fragment of Tir. Our structure revealed that the CesT-binding region in the N-terminus of Tir contains an additional conserved sequence, distinct from the known chaperone-binding β-motif, that we termed the CesT-extension motif because it extends the β-sheet core of CesT. This motif is also present in the C-terminus of Tir that we confirmed to be a unique second CesT-binding region. Point mutations that disrupt CesT-binding to the N- or C-terminus of Tir revealed that the newly identified carboxy-terminal CesT-binding region was required for efficient Tir translocation into HeLa cells and pedestal formation. Furthermore, the CesT-extension motif was identified in the N-terminal region of NleH1, NleH2, and EspZ, and mutations that disrupt this motif reduced translocation of these effectors, and in some cases, overall effector stability, thus validating the universality of this CesT-extension motif. The presence of two CesT-binding regions in Tir, along with the presence of the CesT-extension motif in other highly translocated effectors, may contribute to differential cargo recognition by CesT.
- Department of Biochemistry & Biomedical Sciences, Michael G. DeGroote Institute for Infectious Disease Research, McMaster University, Hamilton, Ontario, Canada.
Organizational Affiliation: