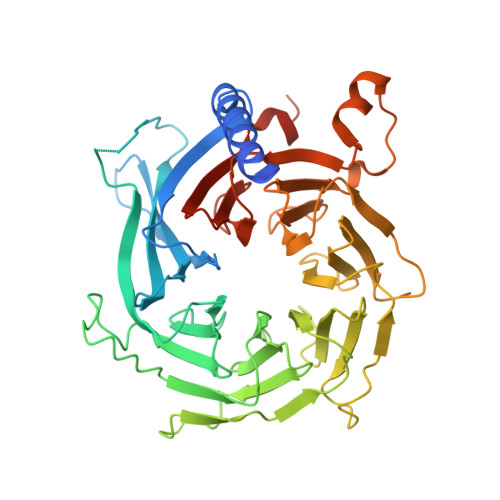
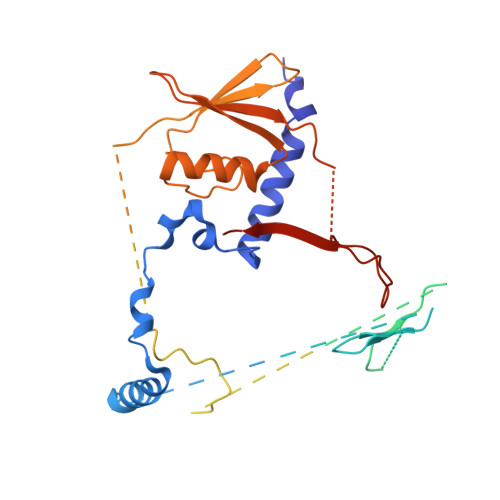
Unique Structural Platforms of Suz12 Dictate Distinct Classes of PRC2 for Chromatin Binding.
Chen, S., Jiao, L., Shubbar, M., Yang, X., Liu, X.(2018) Mol Cell 69: 840-852.e5
- PubMed: 29499137
- DOI: https://doi.org/10.1016/j.molcel.2018.01.039
- Primary Citation of Related Structures:
5WAI, 5WAK - PubMed Abstract:
Developmentally regulated accessory subunits dictate PRC2 function. Here, we report the crystal structures of a 120 kDa heterotetrameric complex consisting of Suz12, Rbbp4, Jarid2, and Aebp2 fragments that is minimally active in nucleosome binding and of an inactive binary complex of Suz12 and Rbbp4. Suz12 contains two unique structural platforms that define distinct classes of PRC2 holo complexes for chromatin binding. Aebp2 and Phf19 compete for binding of a non-canonical C2 domain of Suz12; Jarid2 and EPOP occupy an overlapped Suz12 surface required for chromatin association of PRC2. Suz12 and Aebp2 progressively block histone H3K4 binding to Rbbp4, suggesting that Rbbp4 may not be directly involved in PRC2 inhibition by the active H3K4me3 histone mark. Nucleosome binding enabled by Jarid2 and Aebp2 is in part accounted for by the structures, which also reveal that disruption of the Jarid2-Suz12 interaction may underlie the disease mechanism of an oncogenic chromosomal translocation of Suz12.
- Cecil H. and Ida Green Center for Reproductive Biology Sciences and Division of Basic Research, Department of Obstetrics and Gynecology, Department of Biophysics, UT Southwestern Medical Center, Dallas, TX 75390, USA.
Organizational Affiliation: