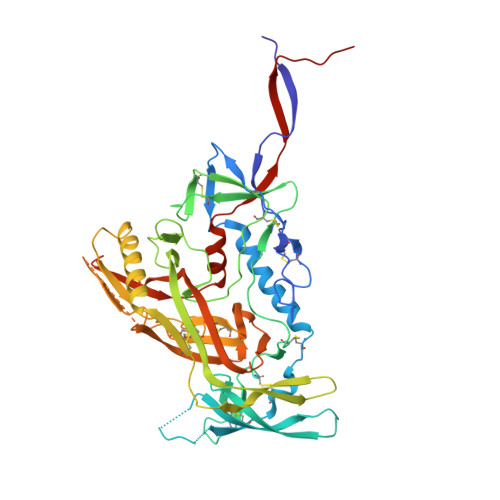
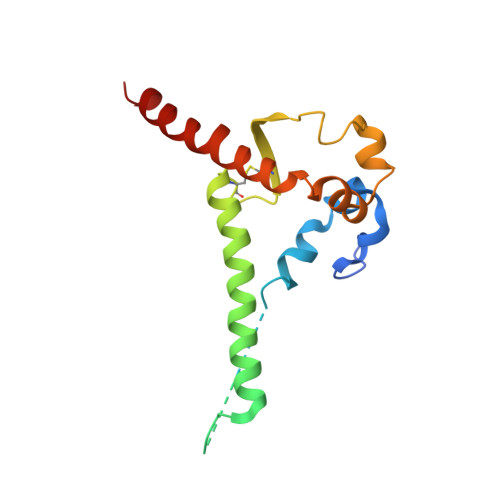
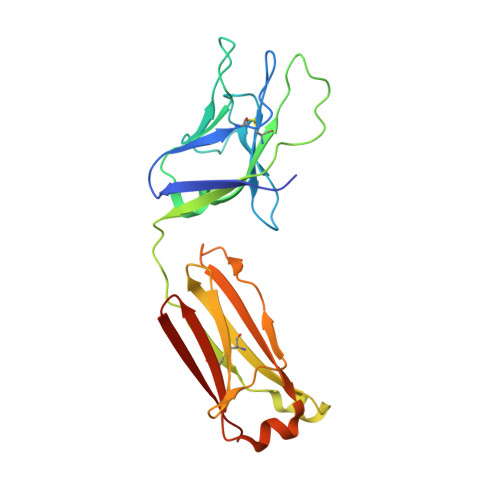
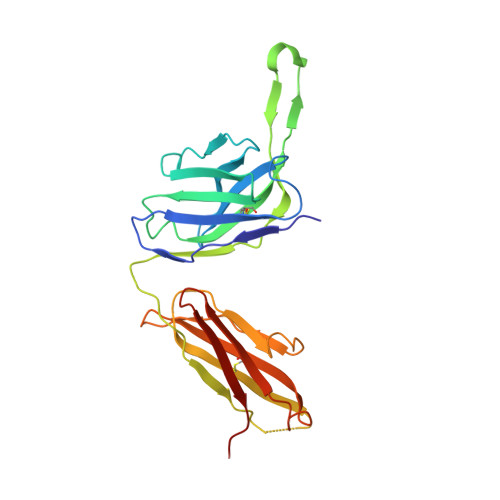
Design and crystal structure of a native-like HIV-1 envelope trimer that engages multiple broadly neutralizing antibody precursors in vivo.
Medina-Ramirez, M., Garces, F., Escolano, A., Skog, P., de Taeye, S.W., Del Moral-Sanchez, I., McGuire, A.T., Yasmeen, A., Behrens, A.J., Ozorowski, G., van den Kerkhof, T.L.G.M., Freund, N.T., Dosenovic, P., Hua, Y., Gitlin, A.D., Cupo, A., van der Woude, P., Golabek, M., Sliepen, K., Blane, T., Kootstra, N., van Breemen, M.J., Pritchard, L.K., Stanfield, R.L., Crispin, M., Ward, A.B., Stamatatos, L., Klasse, P.J., Moore, J.P., Nemazee, D., Nussenzweig, M.C., Wilson, I.A., Sanders, R.W.(2017) J Exp Medicine 214: 2573-2590
- PubMed: 28847869
- DOI: https://doi.org/10.1084/jem.20161160
- Primary Citation of Related Structures:
5W6D - PubMed Abstract:
Induction of broadly neutralizing antibodies (bNAbs) by HIV-1 envelope glycoprotein immunogens would be a major advance toward an effective vaccine. A critical step in this process is the activation of naive B cells expressing germline (gl) antibody precursors that have the potential to evolve into bNAbs. Here, we reengineered the BG505 SOSIP.664 glycoprotein to engage gl precursors of bNAbs that target either the trimer apex or the CD4-binding site. The resulting BG505 SOSIP.v4.1-GT1 trimer binds multiple bNAb gl precursors in vitro. Immunization experiments in knock-in mice expressing gl-VRC01 or gl-PGT121 show that this trimer activates B cells in vivo, resulting in the secretion of specific antibodies into the sera. A crystal structure of the gl-targeting trimer at 3.2-Å resolution in complex with neutralizing antibodies 35O22 and 9H+109L reveals a native-like conformation and the successful incorporation of design features associated with binding of multiple gl-bNAb precursors.
- Department of Medical Microbiology, Academic Medical Center, University of Amsterdam, Amsterdam, Netherlands.
Organizational Affiliation: