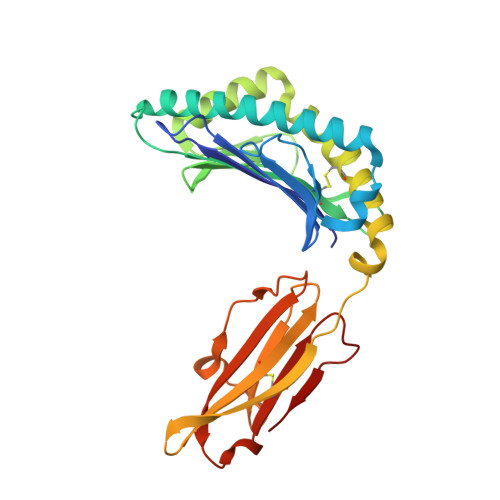
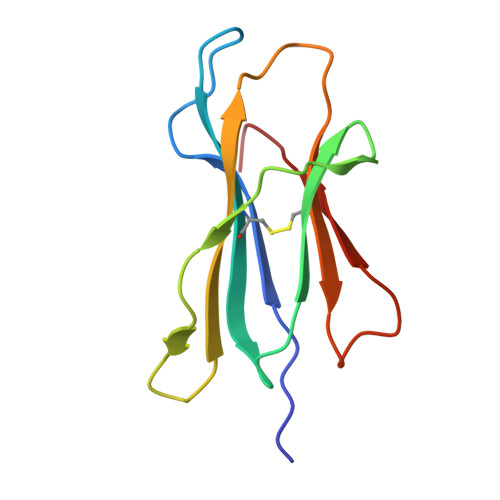
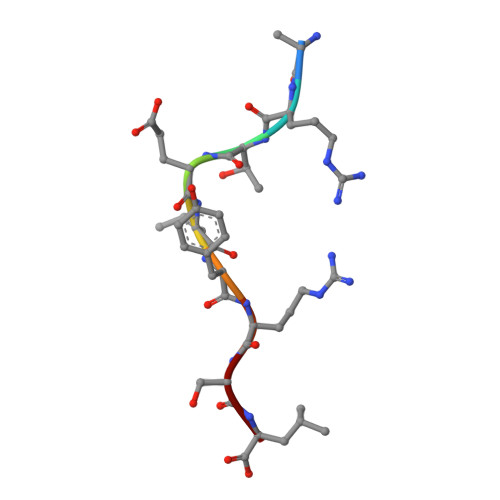
The molecular basis for peptide repertoire selection in the human leucocyte antigen (HLA) C*06:02 molecule.
Mobbs, J.I., Illing, P.T., Dudek, N.L., Brooks, A.G., Baker, D.G., Purcell, A.W., Rossjohn, J., Vivian, J.P.(2017) J Biological Chem 292: 17203-17215
- PubMed: 28855257
- DOI: https://doi.org/10.1074/jbc.M117.806976
- Primary Citation of Related Structures:
5W67, 5W69, 5W6A - PubMed Abstract:
Human leukocyte antigen (HLA)-C*06:02 is identified as the allele associated with the highest risk for the development of the autoimmune skin disease psoriasis. However, the diversity and mode of peptide presentation by the HLA-C*06:02 molecule remains unclear. Here, we describe the endogenous peptide repertoire of ∼3,000 sequences for HLA-C*06:02 that defines the peptide-binding motif for this HLA allomorph. We found that HLA-C*06:02 predominantly presents nonamer peptides with dominant arginine anchors at the P2 and P7 positions and a preference for small hydrophobic residues at the C terminus (PΩ). To determine the structural basis of this selectivity, we determined crystal structures of HLA-C*06:02 in complex with two self-peptides (ARTELYRSL and ARFNDLRFV) and an analogue of a melanocyte autoantigen (ADAMTSL5, VRSRR-abu-LRL) implicated in psoriasis. These structures revealed that HLA-C*06:02 possesses a deep peptide-binding groove comprising two electronegative B- and E-pockets that coincide with the preference for P2 and P7 arginine anchors. The ADAMTSL5 autoantigen possessed a P7-Leu instead of the P7-Arg residue, but nevertheless was accommodated within the HLA-C*06:02 antigen-binding cleft. Collectively, our results provide the structural basis for understanding peptide repertoire selection in HLA-C*06:02.
- From the Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton 3800, Victoria, Australia.
Organizational Affiliation: