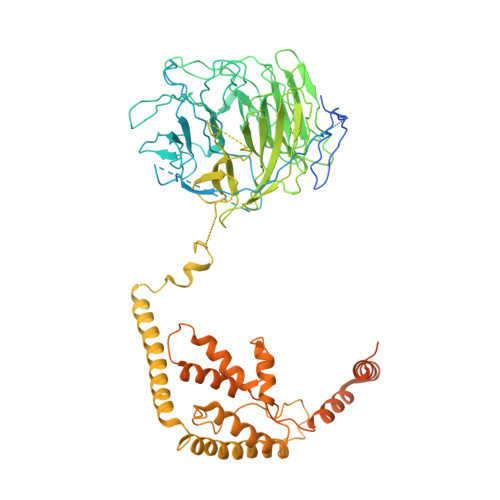
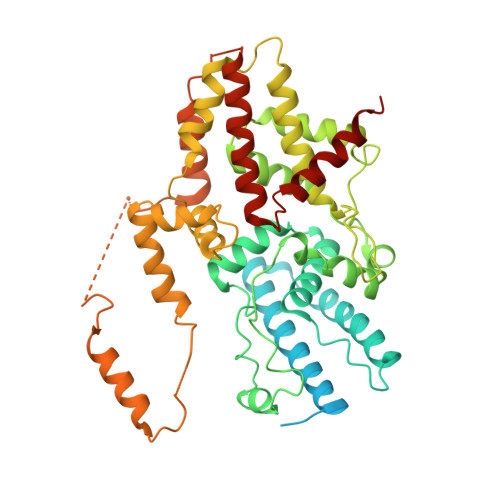
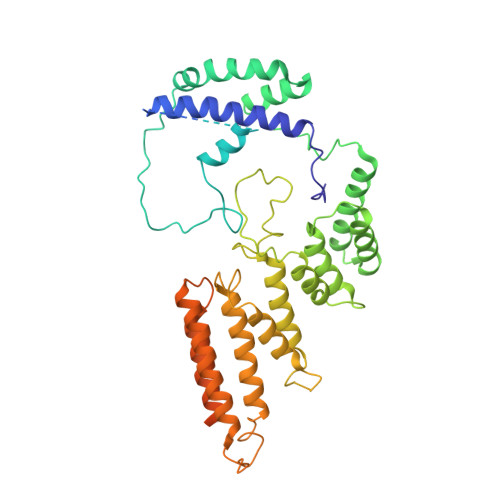
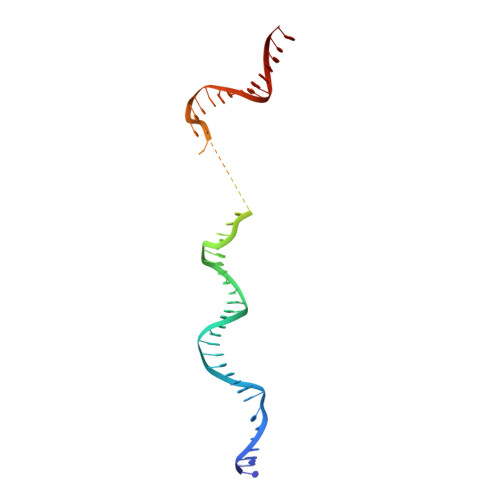
Structural mechanism of ATP-independent transcription initiation by RNA polymerase I.
Han, Y., Yan, C., Nguyen, T.H.D., Jackobel, A.J., Ivanov, I., Knutson, B.A., He, Y.(2017) Elife 6
- PubMed: 28623663
- DOI: https://doi.org/10.7554/eLife.27414
- Primary Citation of Related Structures:
5W5Y, 5W64, 5W65, 5W66 - PubMed Abstract:
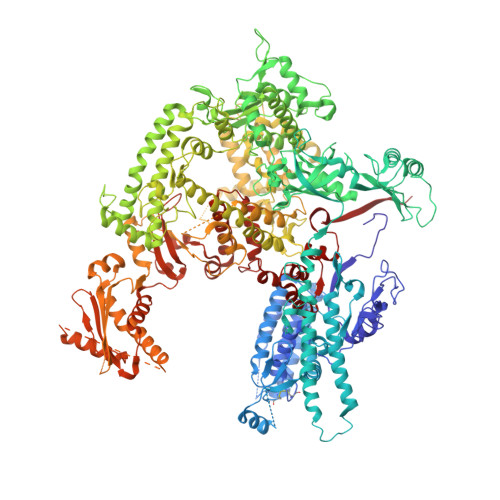
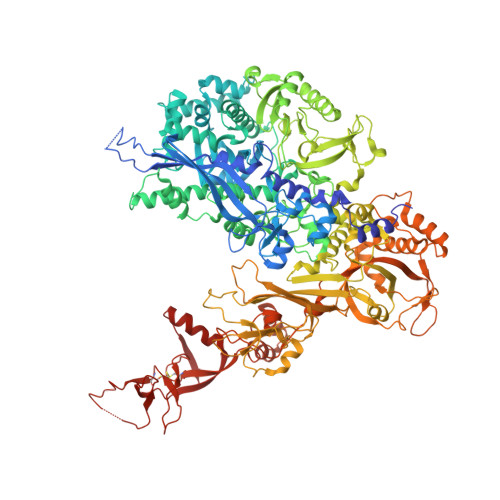
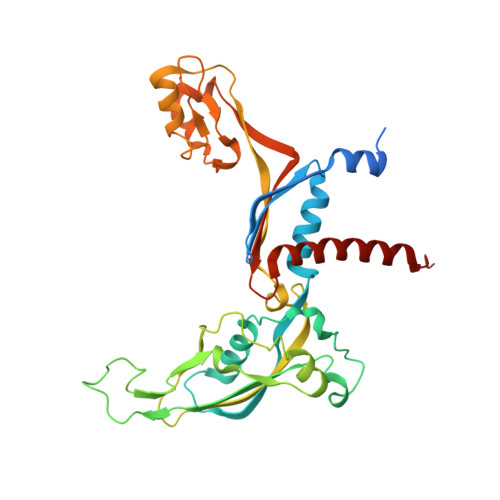
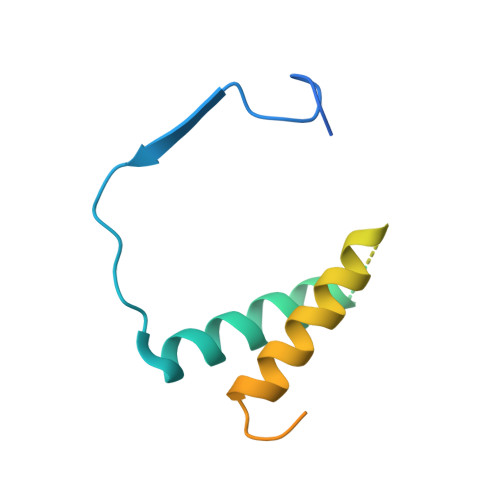
Transcription initiation by RNA Polymerase I (Pol I) depends on the Core Factor (CF) complex to recognize the upstream promoter and assemble into a Pre-Initiation Complex (PIC). Here, we solve a structure of Saccharomyces cerevisiae Pol I-CF-DNA to 3.8 Å resolution using single-particle cryo-electron microscopy. The structure reveals a bipartite architecture of Core Factor and its recognition of the promoter from -27 to -16. Core Factor's intrinsic mobility correlates well with different conformational states of the Pol I cleft, in addition to the stabilization of either Rrn7 N-terminal domain near Pol I wall or the tandem winged helix domain of A49 at a partially overlapping location. Comparison of the three states in this study with the Pol II system suggests that a ratchet motion of the Core Factor-DNA sub-complex at upstream facilitates promoter melting in an ATP-independent manner, distinct from a DNA translocase actively threading the downstream DNA in the Pol II PIC.
- Department of Molecular Biosciences, Northwestern University, Evanston, United States.
Organizational Affiliation: