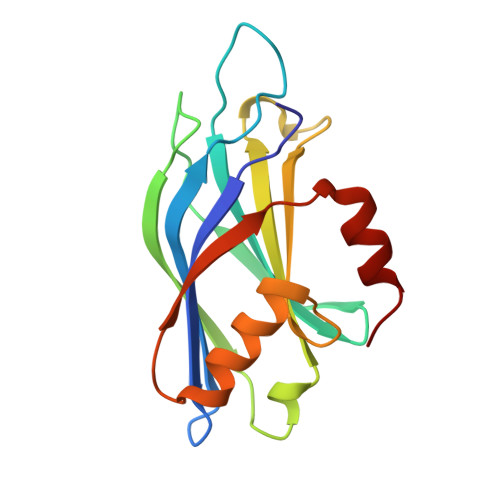
The primed SNARE-complexin-synaptotagmin complex for neuronal exocytosis.
Zhou, Q., Zhou, P., Wang, A.L., Wu, D., Zhao, M., Sudhof, T.C., Brunger, A.T.(2017) Nature 548: 420-425
- PubMed: 28813412
- DOI: https://doi.org/10.1038/nature23484
- Primary Citation of Related Structures:
5W5C, 5W5D - PubMed Abstract:
Synaptotagmin, complexin, and neuronal SNARE (soluble N-ethylmaleimide sensitive factor attachment protein receptor) proteins mediate evoked synchronous neurotransmitter release, but the molecular mechanisms mediating the cooperation between these molecules remain unclear. Here we determine crystal structures of the primed pre-fusion SNARE-complexin-synaptotagmin-1 complex. These structures reveal an unexpected tripartite interface between synaptotagmin-1 and both the SNARE complex and complexin. Simultaneously, a second synaptotagmin-1 molecule interacts with the other side of the SNARE complex via the previously identified primary interface. Mutations that disrupt either interface in solution also severely impair evoked synchronous release in neurons, suggesting that both interfaces are essential for the primed pre-fusion state. Ca 2+ binding to the synaptotagmin-1 molecules unlocks the complex, allows full zippering of the SNARE complex, and triggers membrane fusion. The tripartite SNARE-complexin-synaptotagmin-1 complex at a synaptic vesicle docking site has to be unlocked for triggered fusion to start, explaining the cooperation between complexin and synaptotagmin-1 in synchronizing evoked release on the sub-millisecond timescale.
- Department of Molecular and Cellular Physiology, Howard Hughes Medical Institute, Stanford University, Stanford, California 94305, USA.
Organizational Affiliation: