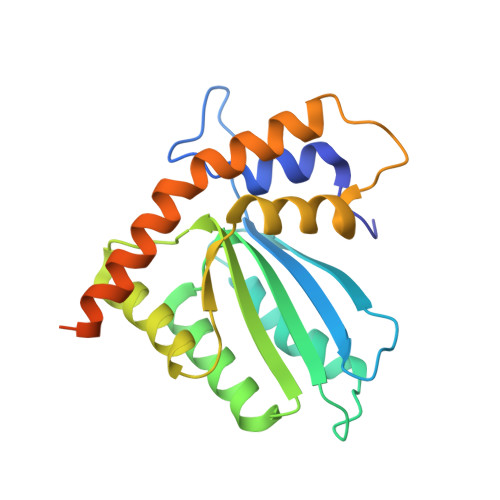
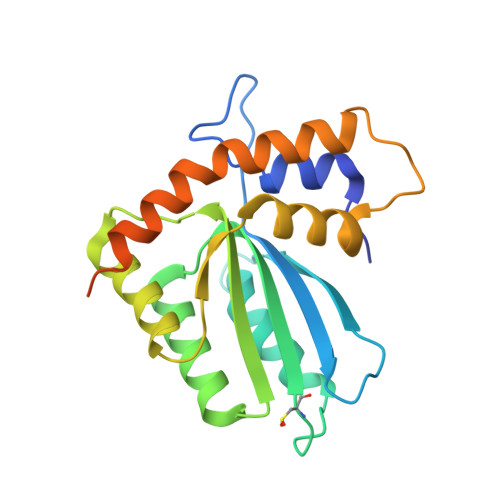
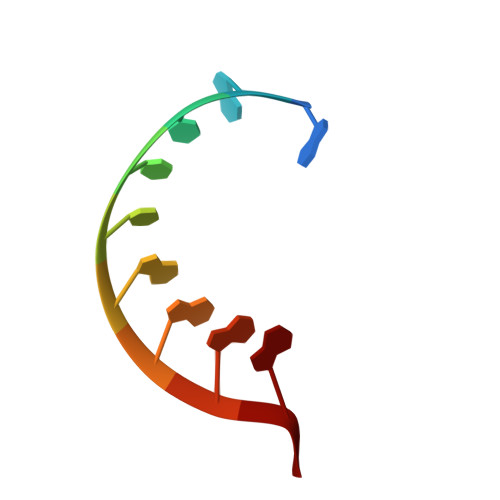
APOBEC3H structure reveals an unusual mechanism of interaction with duplex RNA.
Bohn, J.A., Thummar, K., York, A., Raymond, A., Brown, W.C., Bieniasz, P.D., Hatziioannou, T., Smith, J.L.(2017) Nat Commun 8: 1021-1021
- PubMed: 29044109
- DOI: https://doi.org/10.1038/s41467-017-01309-6
- Primary Citation of Related Structures:
5W3V - PubMed Abstract:
The APOBEC3 family of cytidine deaminases cause lethal hypermutation of retroviruses via deamination of newly reverse-transcribed viral DNA. Their ability to bind RNA is essential for virion infiltration and antiviral activity, yet the mechanisms of viral RNA recognition are unknown. By screening naturally occurring, polymorphic, non-human primate APOBEC3H variants for biological and crystallization properties, we obtained a 2.24-Å crystal structure of pig-tailed macaque APOBEC3H with bound RNA. Here, we report that APOBEC3H forms a dimer around a short RNA duplex and, despite the bound RNA, has potent cytidine deaminase activity. The structure reveals an unusual RNA-binding mode in which two APOBEC3H molecules at opposite ends of a seven-base-pair duplex interact extensively with both RNA strands, but form no protein-protein contacts. CLIP-seq analysis revealed that APOBEC3H preferentially binds to sequences in the viral genome predicted to contain duplexes, a property that may facilitate both virion incorporation and catalytic activity.
- Life Sciences Institute, University of Michigan, Ann Arbor, MI, 48109, USA.
Organizational Affiliation: