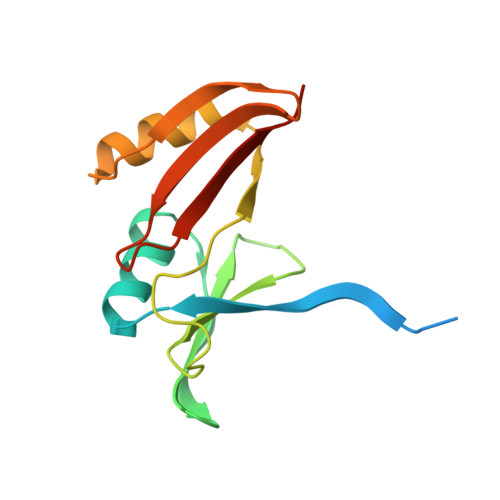
Crystal structure of TnmS3 in complex with tiancimycin (TNM B)
Chang, C.Y., Chang, C., Nocek, B., Rudolf, J.D., Joachimiak, A., Phillips, G.N., SHen, B., Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG)To be published.