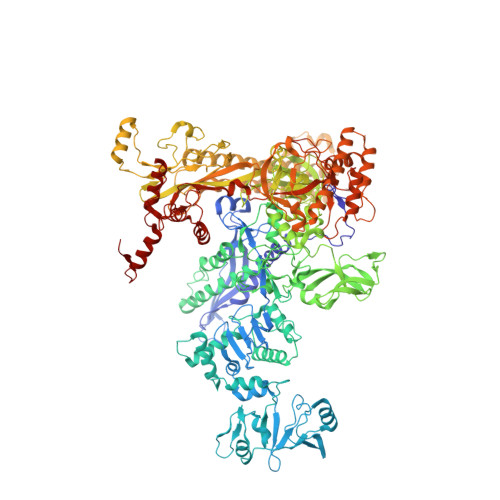
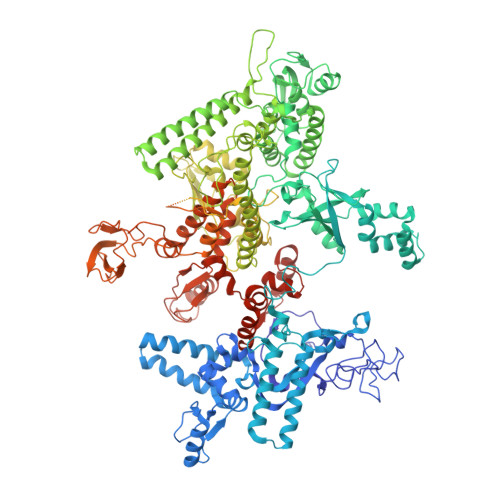
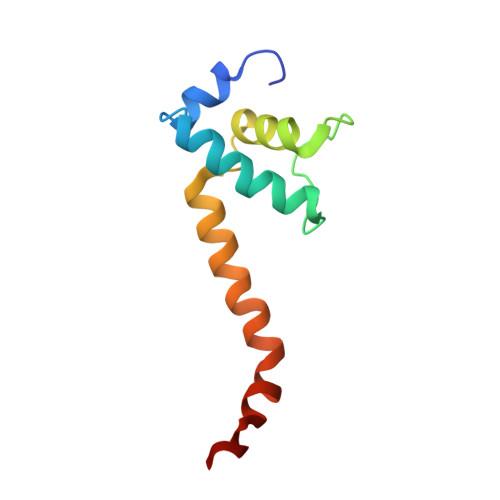
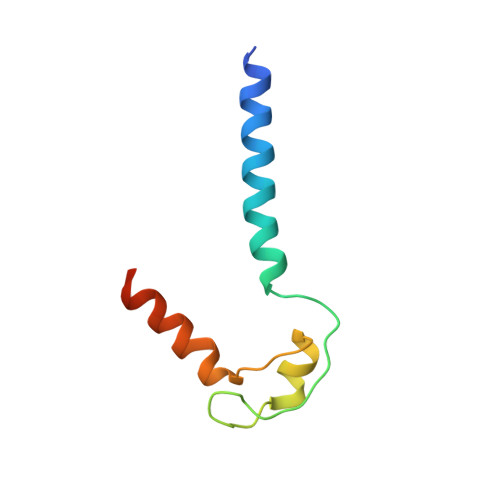
Allosteric Effector ppGpp Potentiates the Inhibition of Transcript Initiation by DksA.
Molodtsov, V., Sineva, E., Zhang, L., Huang, X., Cashel, M., Ades, S.E., Murakami, K.S.(2018) Mol Cell 69: 828-839.e5
- PubMed: 29478808
- DOI: https://doi.org/10.1016/j.molcel.2018.01.035
- Primary Citation of Related Structures:
5VSW, 5W1S, 5W1T - PubMed Abstract:
DksA and ppGpp are the central players in the stringent response and mediate a complete reprogramming of the transcriptome. A major component of the response is a reduction in ribosome synthesis, which is accomplished by the synergistic action of DksA and ppGpp bound to RNA polymerase (RNAP) inhibiting transcription of rRNAs. Here, we report the X-ray crystal structures of Escherichia coli RNAP in complex with DksA alone and with ppGpp. The structures show that DksA accesses the template strand at the active site and the downstream DNA binding site of RNAP simultaneously and reveal that binding of the allosteric effector ppGpp reshapes the RNAP-DksA complex. The structural data support a model for transcriptional inhibition in which ppGpp potentiates the destabilization of open complexes by DksA. This work establishes a structural basis for understanding the pleiotropic effects of DksA and ppGpp on transcriptional regulation in proteobacteria.
- Department of Biochemistry and Molecular Biology, The Center for RNA Molecular Biology, The Pennsylvania State University, University Park, PA 16802, USA.
Organizational Affiliation: