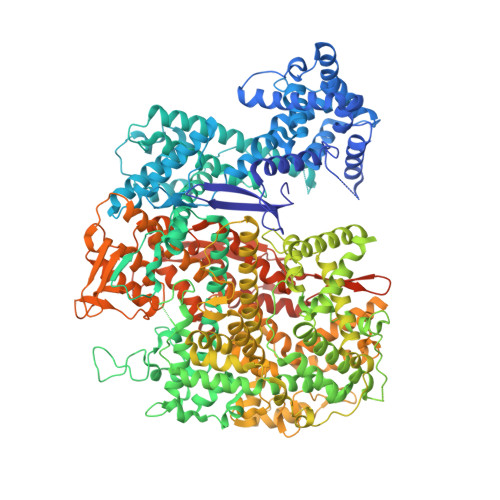
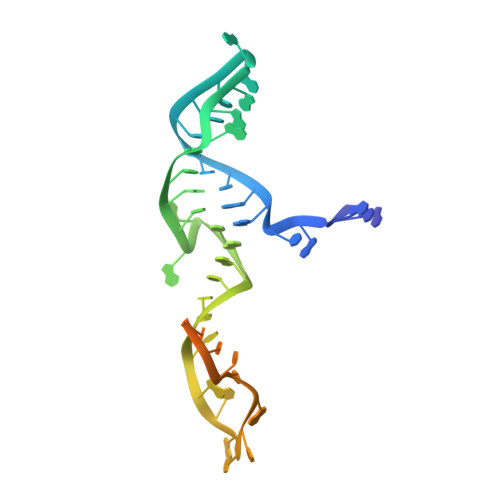
Guide-bound structures of an RNA-targeting A-cleaving CRISPR-Cas13a enzyme.
Knott, G.J., East-Seletsky, A., Cofsky, J.C., Holton, J.M., Charles, E., O'Connell, M.R., Doudna, J.A.(2017) Nat Struct Mol Biol 24: 825-833
- PubMed: 28892041
- DOI: https://doi.org/10.1038/nsmb.3466
- Primary Citation of Related Structures:
5W1H, 5W1I, 5WLH - PubMed Abstract:
CRISPR adaptive immune systems protect bacteria from infections by deploying CRISPR RNA (crRNA)-guided enzymes to recognize and cut foreign nucleic acids. Type VI-A CRISPR-Cas systems include the Cas13a enzyme, an RNA-activated RNase capable of crRNA processing and single-stranded RNA degradation upon target-transcript binding. Here we present the 2.0-Å resolution crystal structure of a crRNA-bound Lachnospiraceae bacterium Cas13a (LbaCas13a), representing a recently discovered Cas13a enzyme subtype. This structure and accompanying biochemical experiments define the Cas13a catalytic residues that are directly responsible for crRNA maturation. In addition, the orientation of the foreign-derived target-RNA-specifying sequence in the protein interior explains the conformational gating of Cas13a nuclease activation. These results describe how Cas13a enzymes generate functional crRNAs and how catalytic activity is blocked before target-RNA recognition, with implications for both bacterial immunity and diagnostic applications.
- Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, California, USA.
Organizational Affiliation: