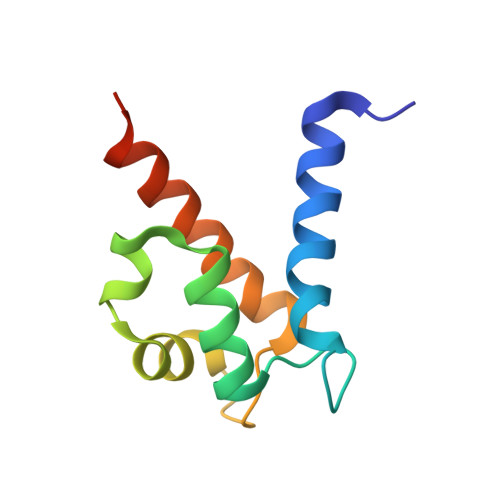
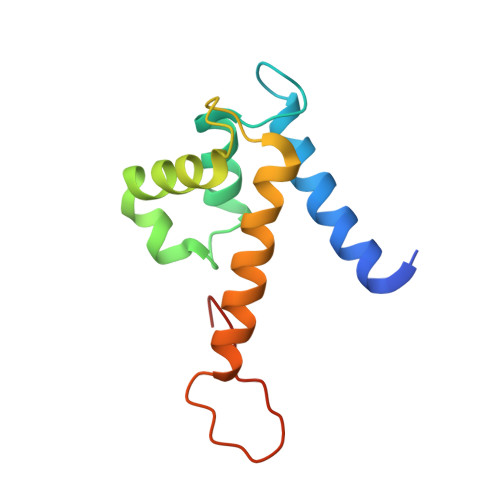
Nickel Sequestration by the Host-Defense Protein Human Calprotectin.
Nakashige, T.G., Zygiel, E.M., Drennan, C.L., Nolan, E.M.(2017) J Am Chem Soc 139: 8828-8836
- PubMed: 28573847
- DOI: https://doi.org/10.1021/jacs.7b01212
- Primary Citation of Related Structures:
5W1F - PubMed Abstract:
The human innate immune protein calprotectin (CP, S100A8/S100A9 oligomer, calgranulin A/calgranulin B oligomer, MRP-8/MRP-14 oligomer) chelates a number of first-row transition metals, including Mn(II), Fe(II), and Zn(II), and can withhold these essential nutrients from microbes. Here we elucidate the Ni(II) coordination chemistry of human CP. We present a 2.6-Å crystal structure of Ni(II)- and Ca(II)-bound CP, which reveals that CP binds Ni(II) ions at both its transition-metal-binding sites: the His 3 Asp motif (site 1) and the His 6 motif (site 2). Further biochemical studies establish that coordination of Ni(II) at the hexahistidine site is thermodynamically preferred over Zn(II). We also demonstrate that CP can sequester Ni(II) from two human pathogens, Staphylococcus aureus and Klebsiella pneumoniae, that utilize this metal nutrient during infection, and inhibit the activity of the Ni(II)-dependent enzyme urease in bacterial cultures. In total, our findings expand the biological coordination chemistry of Ni(II)-chelating proteins in nature and provide a foundation for evaluating putative roles of CP in Ni(II) homeostasis at the host-microbe interface and beyond.
- Department of Chemistry, ‡Department of Biology, and §Howard Hughes Medical Institute, Massachusetts Institute of Technology , Cambridge, Massachusetts 02139, United States.
Organizational Affiliation: