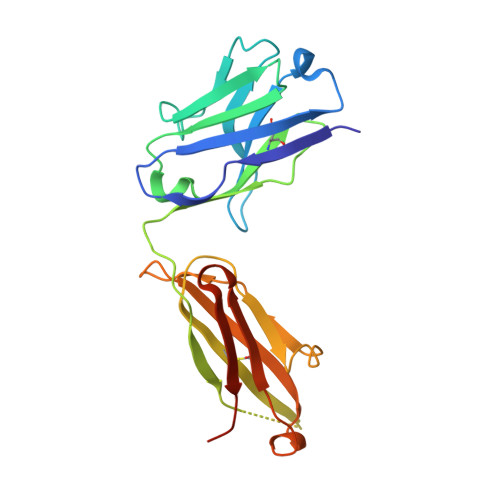
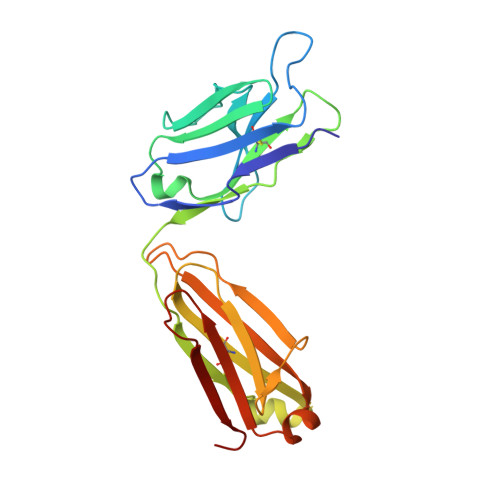
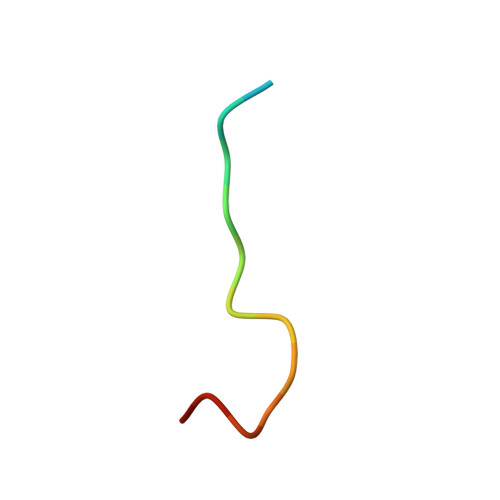Structure of Crenezumab Complex with Abeta Shows Loss of beta-Hairpin.
Ultsch, M., Li, B., Maurer, T., Mathieu, M., Adolfsson, O., Muhs, A., Pfeifer, A., Pihlgren, M., Bainbridge, T.W., Reichelt, M., Ernst, J.A., Eigenbrot, C., Fuh, G., Atwal, J.K., Watts, R.J., Wang, W.(2016) Sci Rep 6: 39374
- PubMed: 27996029
- DOI: https://doi.org/10.1038/srep39374
- Primary Citation of Related Structures:
5VZX, 5VZY - PubMed Abstract:
Accumulation of amyloid-β (Aβ) peptides and amyloid plaque deposition in brain is postulated as a cause of Alzheimer's disease (AD). The precise pathological species of Aβ remains elusive although evidence suggests soluble oligomers may be primarily responsible for neurotoxicity. Crenezumab is a humanized anti-Aβ monoclonal IgG4 that binds multiple forms of Aβ, with higher affinity for aggregated forms, and that blocks Aβ aggregation, and promotes disaggregation. To understand the structural basis for this binding profile and activity, we determined the crystal structure of crenezumab in complex with Aβ. The structure reveals a sequential epitope and conformational requirements for epitope recognition, which include a subtle but critical element that is likely the basis for crenezumab's versatile binding profile. We find interactions consistent with high affinity for multiple forms of Aβ, particularly oligomers. Of note, crenezumab also sequesters the hydrophobic core of Aβ and breaks an essential salt-bridge characteristic of the β-hairpin conformation, eliminating features characteristic of the basic organization in Aβ oligomers and fibrils, and explains crenezumab's inhibition of aggregation and promotion of disaggregation. These insights highlight crenezumab's unique mechanism of action, particularly regarding Aβ oligomers, and provide a strong rationale for the evaluation of crenezumab as a potential AD therapy.
- Genentech, Inc., 1 DNA Way, South San Francisco, California 94080, USA.
Organizational Affiliation: