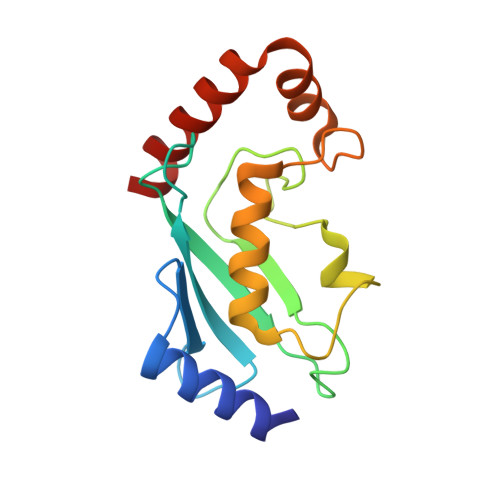
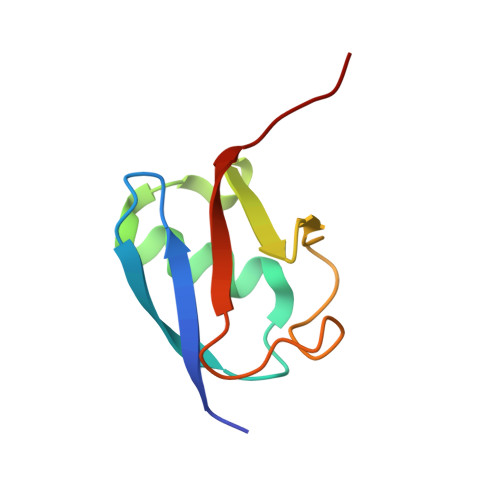
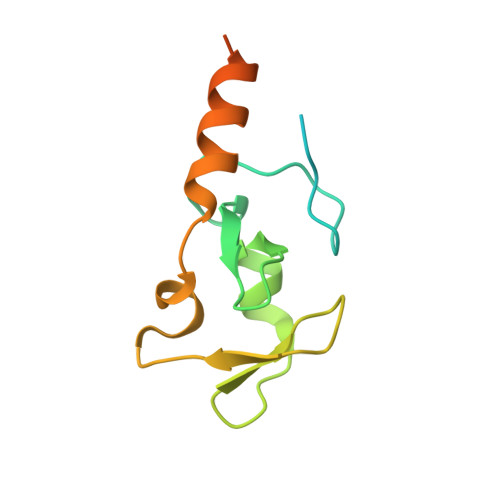
Structure and catalytic activation of the TRIM23 RING E3 ubiquitin ligase.
Dawidziak, D.M., Sanchez, J.G., Wagner, J.M., Ganser-Pornillos, B.K., Pornillos, O.(2017) Proteins 85: 1957-1961
- PubMed: 28681414
- DOI: https://doi.org/10.1002/prot.25348
- Primary Citation of Related Structures:
5VZV, 5VZW - PubMed Abstract:
Tripartite motif (TRIM) proteins comprise a large family of RING-type ubiquitin E3 ligases that regulate important biological processes. An emerging general model is that TRIMs form elongated antiparallel coiled-coil dimers that prevent interaction of the two attendant RING domains. The RING domains themselves bind E2 conjugating enzymes as dimers, implying that an active TRIM ligase requires higher-order oligomerization of the basal coiled-coil dimers. Here, we report crystal structures of the TRIM23 RING domain in isolation and in complex with an E2-ubiquitin conjugate. Our results indicate that TRIM23 enzymatic activity requires RING dimerization, consistent with the general model of TRIM activation.
- Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, Virginia.
Organizational Affiliation: