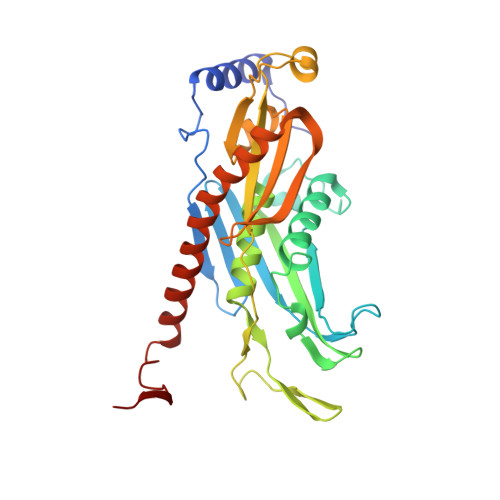
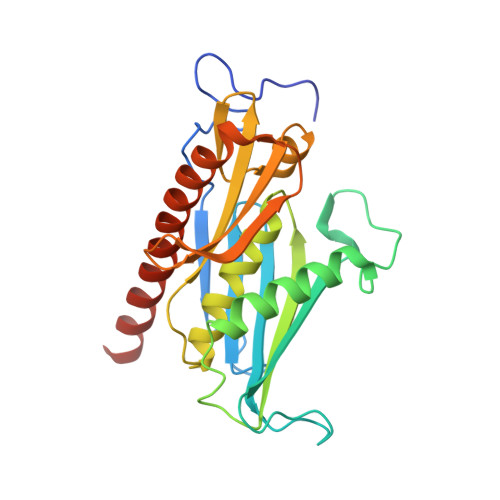
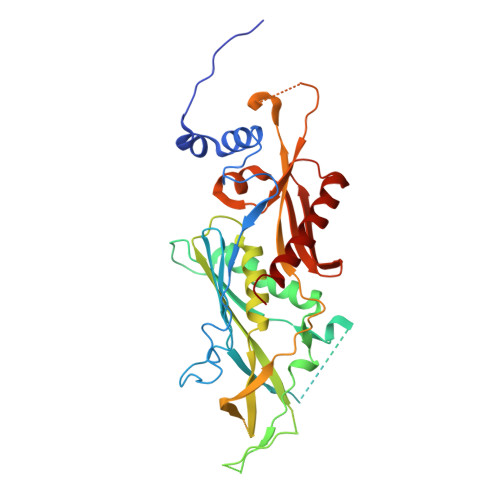
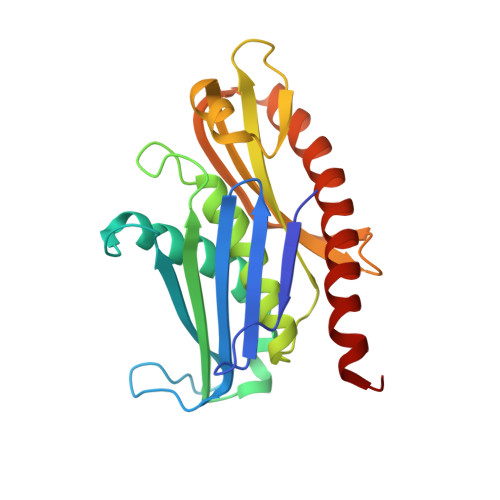
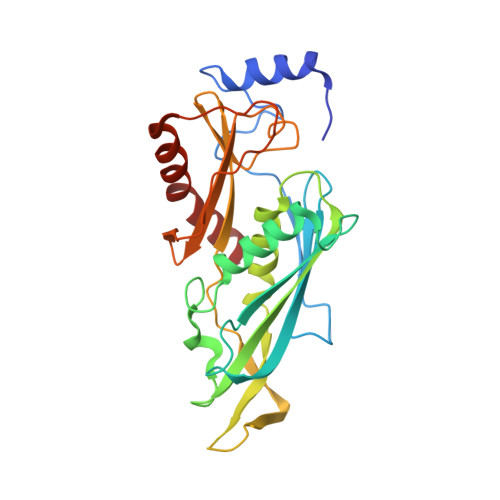
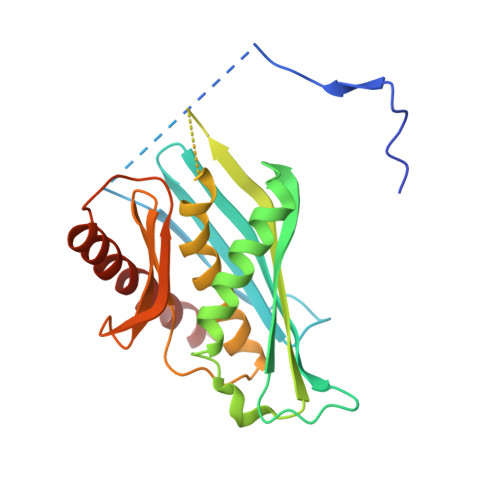
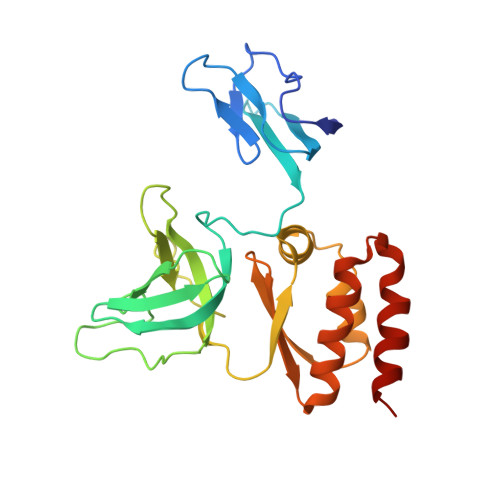
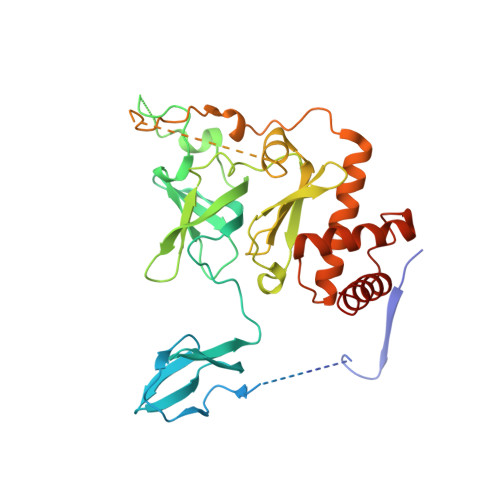
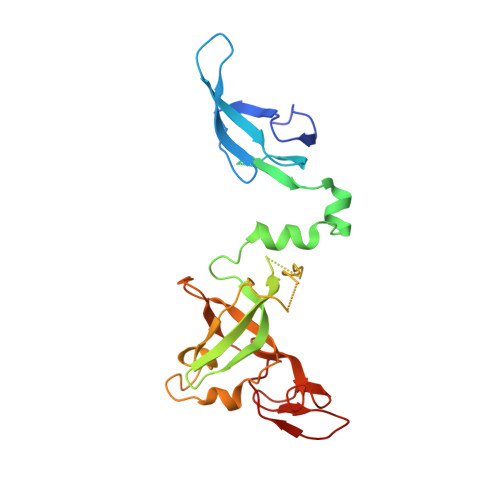
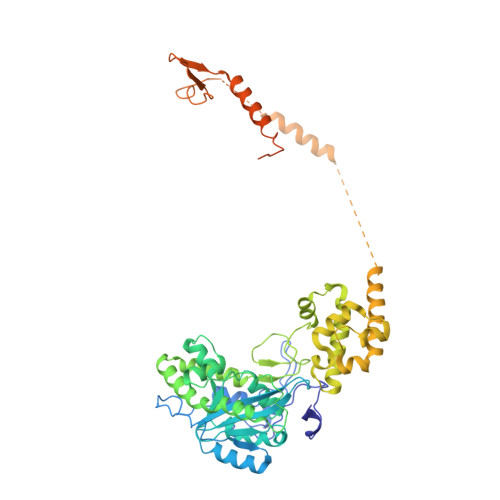
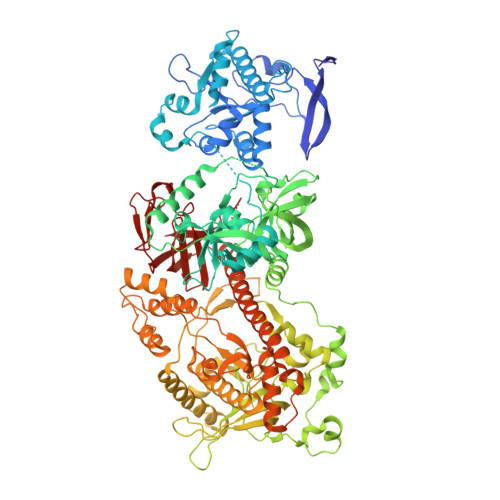
Structure and reconstitution of yeast Mpp6-nuclear exosome complexes reveals that Mpp6 stimulates RNA decay and recruits the Mtr4 helicase.
Wasmuth, E.V., Zinder, J.C., Zattas, D., Das, M., Lima, C.D.(2017) Elife 6
- PubMed: 28742025
- DOI: https://doi.org/10.7554/eLife.29062
- Primary Citation of Related Structures:
5VZJ - PubMed Abstract:
Nuclear RNA exosomes catalyze a range of RNA processing and decay activities that are coordinated in part by cofactors, including Mpp6, Rrp47, and the Mtr4 RNA helicase. Mpp6 interacts with the nine-subunit exosome core, while Rrp47 stabilizes the exoribonuclease Rrp6 and recruits Mtr4, but it is less clear if these cofactors work together. Using biochemistry with Saccharomyces cerevisiae proteins, we show that Rrp47 and Mpp6 stimulate exosome-mediated RNA decay, albeit with unique dependencies on elements within the nuclear exosome. Mpp6-exosomes can recruit Mtr4, while Mpp6 and Rrp47 each contribute to Mtr4-dependent RNA decay, with maximal Mtr4-dependent decay observed with both cofactors. The 3.3 Å structure of a twelve-subunit nuclear Mpp6 exosome bound to RNA shows the central region of Mpp6 bound to the exosome core, positioning its Mtr4 recruitment domain next to Rrp6 and the exosome central channel. Genetic analysis reveals interactions that are largely consistent with our model.
- Structural Biology Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, United States.
Organizational Affiliation: