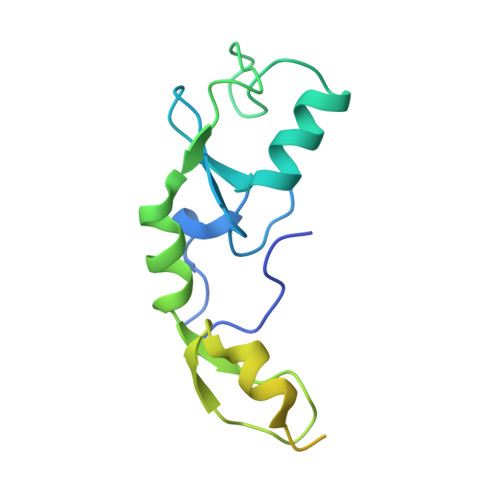
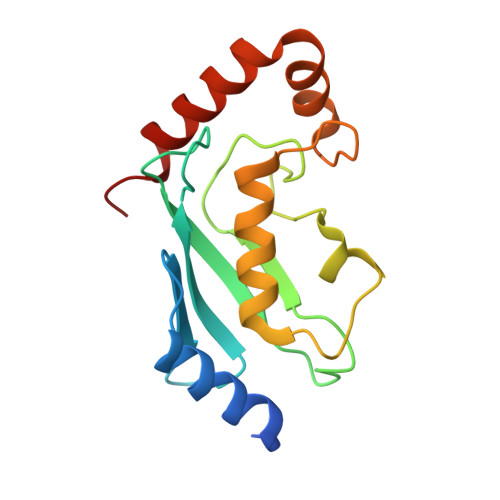
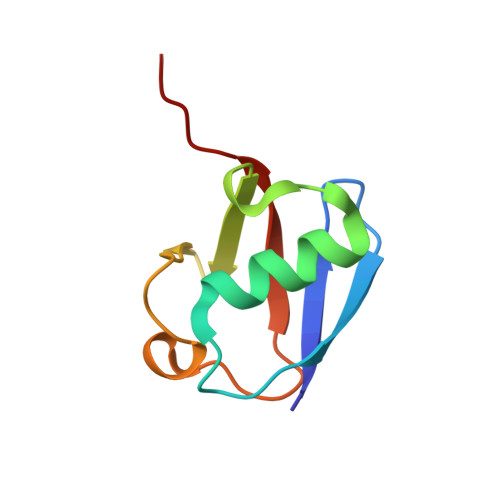
The activity of TRAF RING homo- and heterodimers is regulated by zinc finger 1.
Middleton, A.J., Budhidarmo, R., Das, A., Zhu, J., Foglizzo, M., Mace, P.D., Day, C.L.(2017) Nat Commun 8: 1788-1788
- PubMed: 29176576
- DOI: https://doi.org/10.1038/s41467-017-01665-3
- Primary Citation of Related Structures:
5VNZ, 5VO0 - PubMed Abstract:
Ubiquitin chains linked through lysine63 (K63) play a critical role in inflammatory signalling. Following ligand engagement of immune receptors, the RING E3 ligase TRAF6 builds K63-linked chains together with the heterodimeric E2 enzyme Ubc13-Uev1A. Dimerisation of the TRAF6 RING domain is essential for the assembly of K63-linked ubiquitin chains. Here, we show that TRAF6 RING dimers form a catalytic complex where one RING interacts with a Ubc13~Ubiquitin conjugate, while the zinc finger 1 (ZF1) domain and linker-helix of the opposing monomer contact ubiquitin. The RING dimer interface is conserved across TRAFs and we also show that TRAF5-TRAF6 heterodimers form. Importantly, TRAF5 can provide ZF1, enabling ubiquitin transfer from a TRAF6-bound Ubc13 conjugate. Our study explains the dependence of activity on TRAF RING dimers, and suggests that both homo- and heterodimers mediated by TRAF RING domains have the capacity to synthesise ubiquitin chains.
- Department of Biochemistry, School of Biomedical Sciences, University of Otago, Dunedin, 9054, New Zealand.
Organizational Affiliation: