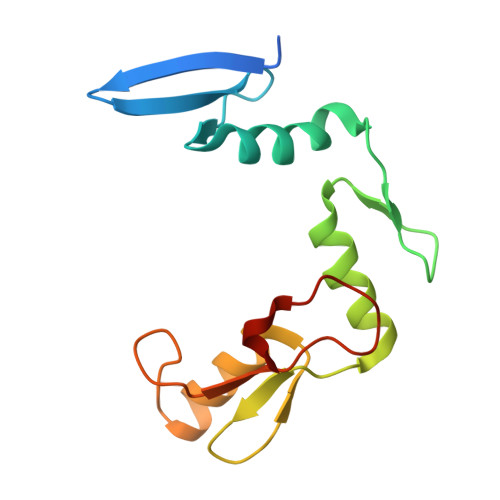
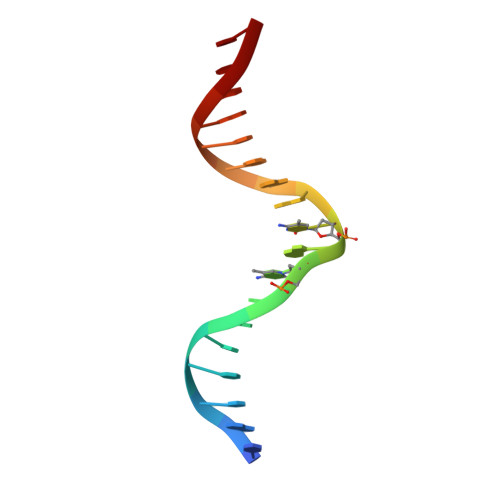
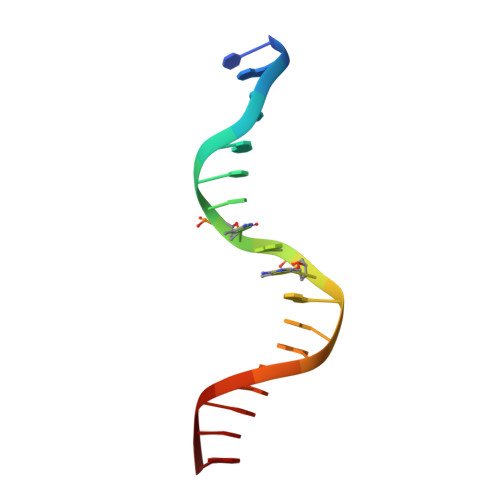
CH···O Hydrogen Bonds Mediate Highly Specific Recognition of Methylated CpG Sites by the Zinc Finger Protein Kaiso.
Nikolova, E.N., Stanfield, R.L., Dyson, H.J., Wright, P.E.(2018) Biochemistry 57: 2109-2120
- PubMed: 29546986
- DOI: https://doi.org/10.1021/acs.biochem.8b00065
- Primary Citation of Related Structures:
5VMU, 5VMV, 5VMW, 5VMX, 5VMY, 5VMZ - PubMed Abstract:
Many eukaryotic transcription factors recognize the epigenetic marker 5-methylcytosine (mC) at CpG sites in DNA. Despite their structural diversity, methyl-CpG-binding proteins (MBPs) share a common mode of recognition of mC methyl groups that involves hydrophobic pockets and weak hydrogen bonds of the CH···O type. The zinc finger protein Kaiso possesses a remarkably high specificity for methylated over unmethylated CpG sites. A key contribution to this specificity is provided by glutamate 535 (E535), which is optimally positioned to form multiple interactions with mCpG, including direct CH···O hydrogen bonds. To examine the role of E535 and CH···O hydrogen bonding in the preferential recognition of mCpG sites, we determined the structures of wild type Kaiso (WT) and E535 mutants and characterized their interactions with methylated DNA by nuclear magnetic resonance spectroscopy (NMR), X-ray crystallography, and in vitro protein-DNA binding assays. Our data show that Kaiso favors an mCpG over a CpG site by 2 orders of magnitude in affinity and that an important component of this effect is the presence of hydrophobic and CH···O contacts involving E535. Moreover, we present the first direct evidence for formation of a CH···O hydrogen bond between an MBP and 5-methylcytosine by using experimental (NMR) and quantum mechanical chemical shift analysis of the mC methyl protons. Together, our findings uncover a critical function of methyl-specific interactions, including CH···O hydrogen bonds, that optimize the specificity and affinity of MBPs for methylated DNA and contribute to the precise control of gene expression.
- Department of Integrative Structural and Computational Biology and Skaggs Institute of Chemical Biology , The Scripps Research Institute , 10550 North Torrey Pines Road , La Jolla , California 92037 , United States.
Organizational Affiliation: