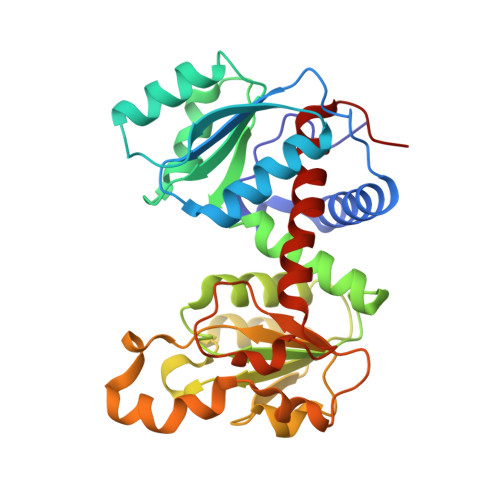
Charge neutralization in the active site of the catalytic trimer of aspartate transcarbamoylase promotes diverse structural changes.
Endrizzi, J.A., Beernink, P.T.(2017) Protein Sci 26: 2221-2228
- PubMed: 28833948
- DOI: https://doi.org/10.1002/pro.3277
- Primary Citation of Related Structures:
5VMQ - PubMed Abstract:
A classical model for allosteric regulation of enzyme activity posits an equilibrium between inactive and active conformations. An alternative view is that allosteric activation is achieved by increasing the potential for conformational changes that are essential for catalysis. In the present study, substitution of a basic residue in the active site of the catalytic (C) trimer of aspartate transcarbamoylase with a non-polar residue results in large interdomain hinge changes in the three chains of the trimer. One conformation is more open than the chains in both the wild-type C trimer and the catalytic chains in the holoenzyme, the second is closed similar to the bisubstrate-analog bound conformation and the third hinge angle is intermediate to the other two. The active-site 240s loop conformation is very different between the most open and closed chains, and is disordered in the third chain, as in the holoenzyme. We hypothesize that binding of anionic substrates may promote similar structural changes. Further, the ability of the three catalytic chains in the trimer to access the open and closed active-site conformations simultaneously suggests a cyclic catalytic mechanism, in which at least one of the chains is in an open conformation suitable for substrate binding whereas another chain is closed for catalytic turnover. Based on the many conformations observed for the chains in the isolated catalytic trimer to date, we propose that allosteric activation of the holoenzyme occurs by release of quaternary constraint into an ensemble of active-site conformations.
- Supernova C, Missoula, Montana.
Organizational Affiliation: