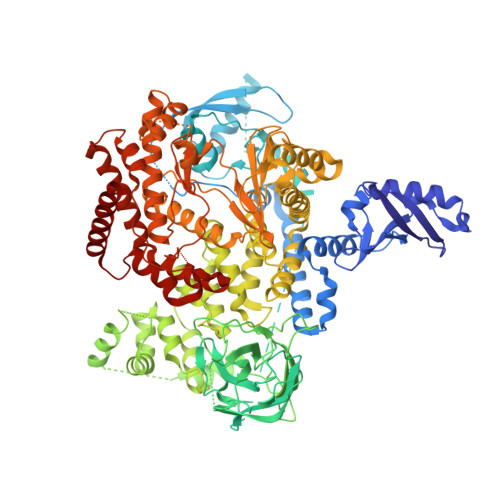
Identification of a Potent, Selective, and Efficacious Phosphatidylinositol 3-Kinase delta (PI3K delta ) Inhibitor for the Treatment of Immunological Disorders.
Liu, Q., Shi, Q., Marcoux, D., Batt, D.G., Cornelius, L., Qin, L.Y., Ruan, Z., Neels, J., Beaudoin-Bertrand, M., Srivastava, A.S., Li, L., Cherney, R.J., Gong, H., Watterson, S.H., Weigelt, C., Gillooly, K.M., McIntyre, K.W., Xie, J.H., Obermeier, M.T., Fura, A., Sleczka, B., Stefanski, K., Fancher, R.M., Padmanabhan, S., Rp, T., Kundu, I., Rajareddy, K., Smith, R., Hennan, J.K., Xing, D., Fan, J., Levesque, P.C., Ruan, Q., Pitt, S., Zhang, R., Pedicord, D., Pan, J., Yarde, M., Lu, H., Lippy, J., Goldstine, C., Skala, S., Rampulla, R.A., Mathur, A., Gupta, A., Arunachalam, P.N., Sack, J.S., Muckelbauer, J.K., Cvijic, M.E., Salter-Cid, L.M., Bhide, R.S., Poss, M.A., Hynes, J., Carter, P.H., Macor, J.E., Ruepp, S., Schieven, G.L., Tino, J.A.(2017) J Med Chem 60: 5193-5208
- PubMed: 28541707
- DOI: https://doi.org/10.1021/acs.jmedchem.7b00618
- Primary Citation of Related Structures:
5VLR - PubMed Abstract:
PI3Kδ plays an important role controlling immune cell function and has therefore been identified as a potential target for the treatment of immunological disorders. This article highlights our work toward the identification of a potent, selective, and efficacious PI3Kδ inhibitor. Through careful SAR, the successful replacement of a polar pyrazole group by a simple chloro or trifluoromethyl group led to improved Caco-2 permeability, reduced Caco-2 efflux, reduced hERG PC activity, and increased selectivity profile while maintaining potency in the CD69 hWB assay. The optimization of the aryl substitution then identified a 4'-CN group that improved the human/rodent correlation in microsomal metabolic stability. Our lead molecule is very potent in PK/PD assays and highly efficacious in a mouse collagen-induced arthritis model.
- Research & Development, Bristol-Myers Squibb Company , Route 206 and Province Line Road, Princeton, New Jersey 08543, United States.
Organizational Affiliation: