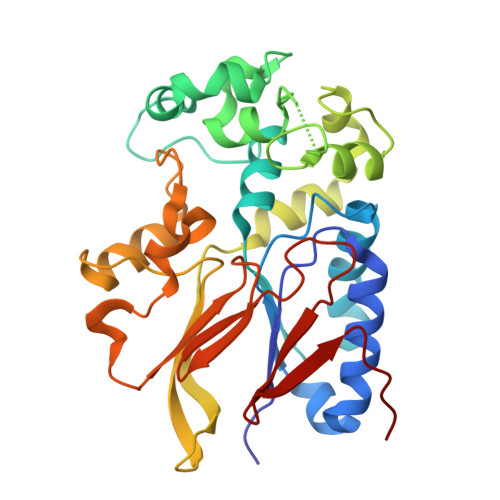
Structural basis for the preference of the Arabidopsis thalianaphosphatase RLPH2 for tyrosine-phosphorylated substrates.
Labandera, A.M., Uhrig, R.G., Colville, K., Moorhead, G.B., Ng, K.K.S.(2018) Sci Signal 11
- PubMed: 29615518
- DOI: https://doi.org/10.1126/scisignal.aan8804
- Primary Citation of Related Structures:
5VJV, 5VJW - PubMed Abstract:
Despite belonging to the phosphoserine- and phosphothreonine-specific phosphoprotein phosphatase (PPP) family, Arabidopsis thaliana Rhizobiales -like phosphatase 2 (RLPH2) strongly prefers substrates bearing phosphorylated tyrosine residues. We solved the structures of RLPH2 crystallized in the presence or absence of sodium tungstate. These structures revealed the presence of a central domain that forms a binding site for two divalent metal ions that closely resembles that of other PPP-family enzymes. Unique structural elements from two flanking domains suggest a mechanism for the selective dephosphorylation of phosphotyrosine residues. Cocrystallization with the phosphate mimetic tungstate also suggests how positively charged residues that are highly conserved in the RLPH2 class form an additional pocket that is specific for a phosphothreonine residue located near the phosphotyrosine residue that is bound to the active site. Site-directed mutagenesis confirmed that this auxiliary recognition element facilitates the recruitment of dual-phosphorylated substrates containing a pTxpY motif.
- Department of Biological Sciences, University of Calgary, 2500 University Drive Northwest, Calgary, Alberta T2N 1N4, Canada.
Organizational Affiliation: