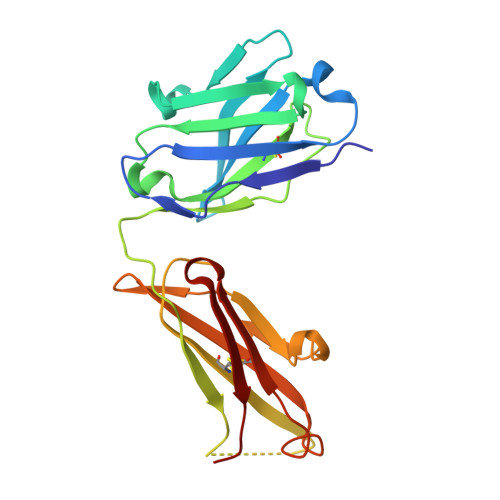
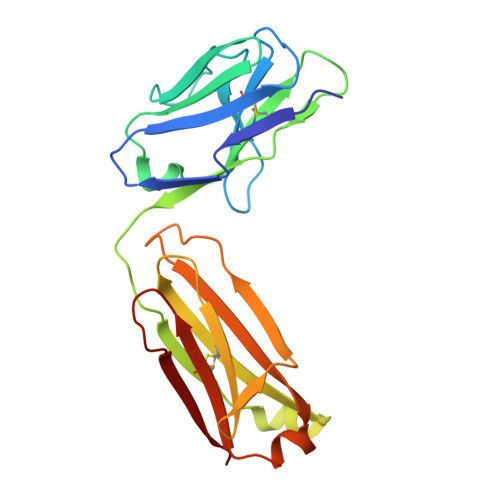
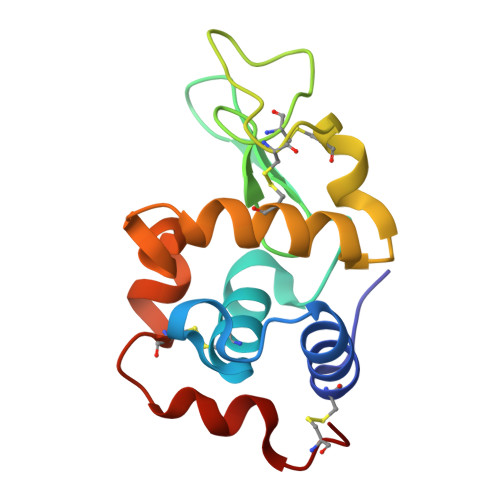
Germinal center antibody mutation trajectories are determined by rapid self/foreign discrimination.
Burnett, D.L., Langley, D.B., Schofield, P., Hermes, J.R., Chan, T.D., Jackson, J., Bourne, K., Reed, J.H., Patterson, K., Porebski, B.T., Brink, R., Christ, D., Goodnow, C.C.(2018) Science 360: 223-226
- PubMed: 29650674
- DOI: https://doi.org/10.1126/science.aao3859
- Primary Citation of Related Structures:
5VJO, 5VJQ - PubMed Abstract:
Antibodies have the specificity to differentiate foreign antigens that mimic self antigens, but it remains unclear how such specificity is acquired. In a mouse model, we generated B cells displaying an antibody that cross-reacts with two related protein antigens expressed on self versus foreign cells. B cell anergy was imposed by self antigen but reversed upon challenge with high-density foreign antigen, leading to germinal center recruitment and antibody gene hypermutation. Single-cell analysis detected rapid selection for mutations that decrease self affinity and slower selection for epistatic mutations that specifically increase foreign affinity. Crystal structures revealed that these mutations exploited subtle topological differences to achieve 5000-fold preferential binding to foreign over self epitopes. Resolution of antigenic mimicry drove the optimal affinity maturation trajectory, highlighting the value of retaining self-reactive clones as substrates for protective antibody responses.
- Garvan Institute of Medical Research, 384 Victoria Street, Darlinghurst, New South Wales 2010, Australia.
Organizational Affiliation: