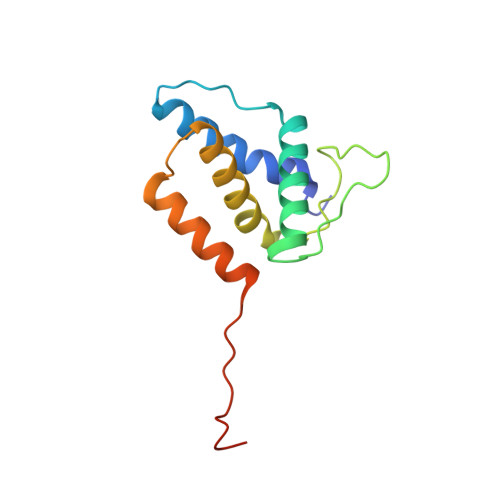
High-resolution structure of podovirus tail adaptor suggests repositioning of an octad motif that mediates the sequential tail assembly.
Liang, L., Zhao, H., An, B., Tang, L.(2018) Proc Natl Acad Sci U S A 115: 313-318
- PubMed: 29279385
- DOI: https://doi.org/10.1073/pnas.1706846115
- Primary Citation of Related Structures:
5VGT - PubMed Abstract:
The sophisticated tail structures of DNA bacteriophages play essential roles in life cycles. Podoviruses P22 and Sf6 have short tails consisting of multiple proteins, among which is a tail adaptor protein that connects the portal protein to the other tail proteins. Assembly of the tail has been shown to occur in a sequential manner to ensure proper molecular interactions, but the underlying mechanism remains to be understood. Here, we report the high-resolution structure of the tail adaptor protein gp7 from phage Sf6. The structure exhibits distinct distribution of opposite charges on two sides of the molecule. A gp7 dodecameric ring model shows an entirely negatively charged surface, suggesting that the assembly of the dodecamer occurs through head-to-tail interactions of the bipolar monomers. The N-terminal helix-loop structure undergoes rearrangement compared with that of the P22 homolog complexed with the portal, which is achieved by repositioning of two consecutive repeats of a conserved octad sequence motif. We propose that the conformation of the N-terminal helix-loop observed in the Sf6-gp7 and P22 portal:gp4 complex represents the pre- and postassembly state, respectively. Such motif repositioning may serve as a conformational switch that creates the docking site for the tail nozzle only after the assembly of adaptor protein to the portal. In addition, the C-terminal portion of gp7 shows conformational flexibility, indicating an induced fit on binding to the portal. These results provide insight into the mechanistic role of the adaptor protein in mediating the sequential assembly of the phage tail.
- Department of Molecular Biosciences, University of Kansas, Lawrence, KS 66045.
Organizational Affiliation: