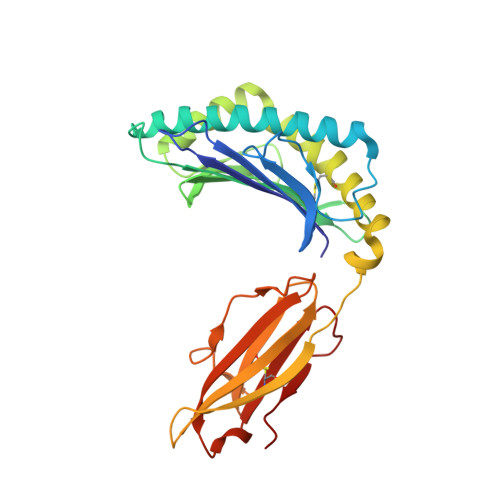
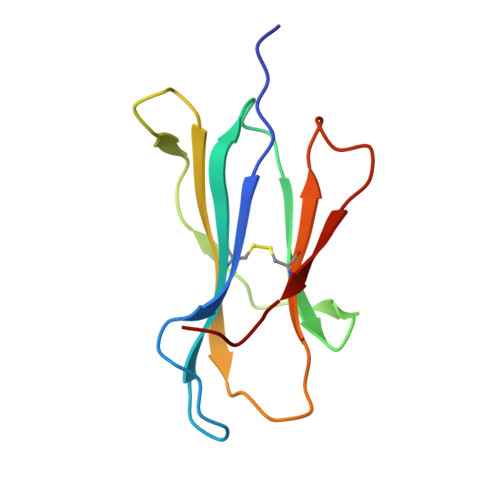
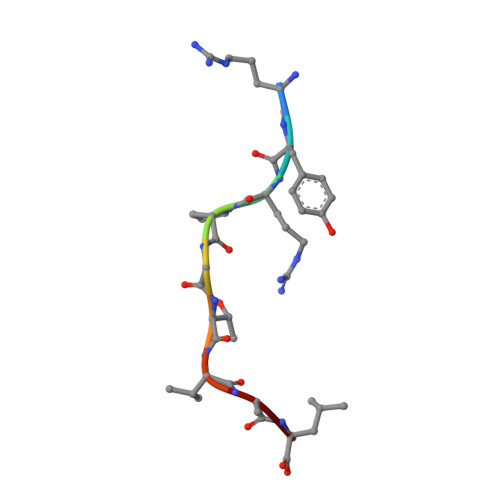
Structural and regulatory diversity shape HLA-C protein expression levels.
Kaur, G., Gras, S., Mobbs, J.I., Vivian, J.P., Cortes, A., Barber, T., Kuttikkatte, S.B., Jensen, L.T., Attfield, K.E., Dendrou, C.A., Carrington, M., McVean, G., Purcell, A.W., Rossjohn, J., Fugger, L.(2017) Nat Commun 8: 15924-15924
- PubMed: 28649982
- DOI: https://doi.org/10.1038/ncomms15924
- Primary Citation of Related Structures:
5VGD, 5VGE - PubMed Abstract:
Expression of HLA-C varies widely across individuals in an allele-specific manner. This variation in expression can influence efficacy of the immune response, as shown for infectious and autoimmune diseases. MicroRNA binding partially influences differential HLA-C expression, but the additional contributing factors have remained undetermined. Here we use functional and structural analyses to demonstrate that HLA-C expression is modulated not just at the RNA level, but also at the protein level. Specifically, we show that variation in exons 2 and 3, which encode the α1/α2 domains, drives differential expression of HLA-C allomorphs at the cell surface by influencing the structure of the peptide-binding cleft and the diversity of peptides bound by the HLA-C molecules. Together with a phylogenetic analysis, these results highlight the diversity and long-term balancing selection of regulatory factors that modulate HLA-C expression.
- MRC Human Immunology Unit, Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, University of Oxford, Oxford OX3 9DS, UK.
Organizational Affiliation: