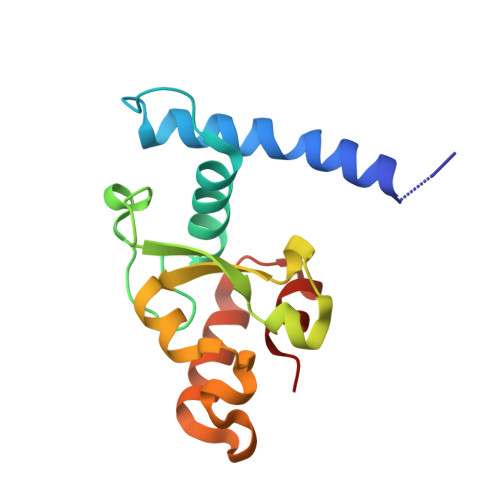
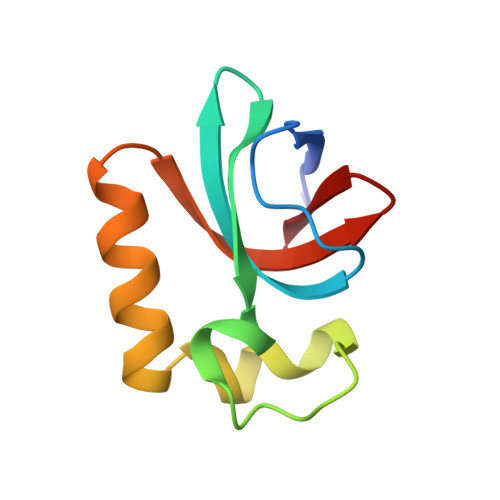
A Broad-Spectrum Inhibitor of CRISPR-Cas9.
Harrington, L.B., Doxzen, K.W., Ma, E., Liu, J.J., Knott, G.J., Edraki, A., Garcia, B., Amrani, N., Chen, J.S., Cofsky, J.C., Kranzusch, P.J., Sontheimer, E.J., Davidson, A.R., Maxwell, K.L., Doudna, J.A.(2017) Cell 170: 1224-1233.e15
- PubMed: 28844692
- DOI: https://doi.org/10.1016/j.cell.2017.07.037
- Primary Citation of Related Structures:
5VGB - PubMed Abstract:
CRISPR-Cas9 proteins function within bacterial immune systems to target and destroy invasive DNA and have been harnessed as a robust technology for genome editing. Small bacteriophage-encoded anti-CRISPR proteins (Acrs) can inactivate Cas9, providing an efficient off switch for Cas9-based applications. Here, we show that two Acrs, AcrIIC1 and AcrIIC3, inhibit Cas9 by distinct strategies. AcrIIC1 is a broad-spectrum Cas9 inhibitor that prevents DNA cutting by multiple divergent Cas9 orthologs through direct binding to the conserved HNH catalytic domain of Cas9. A crystal structure of an AcrIIC1-Cas9 HNH domain complex shows how AcrIIC1 traps Cas9 in a DNA-bound but catalytically inactive state. By contrast, AcrIIC3 blocks activity of a single Cas9 ortholog and induces Cas9 dimerization while preventing binding to the target DNA. These two orthogonal mechanisms allow for separate control of Cas9 target binding and cleavage and suggest applications to allow DNA binding while preventing DNA cutting by Cas9.
- Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, CA 94720, USA.
Organizational Affiliation: