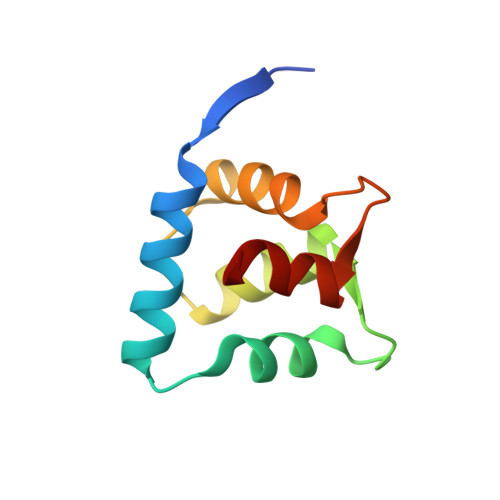
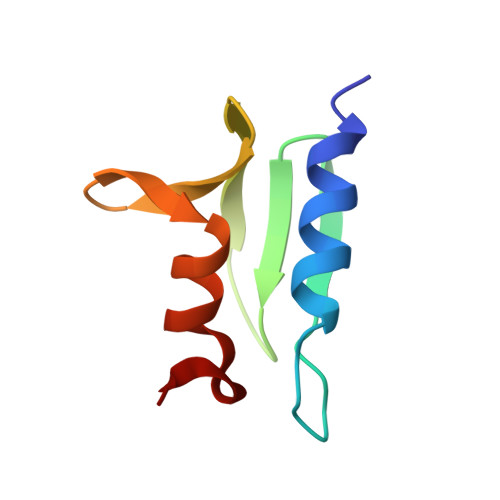
Structure of the ACF7 EF-Hand-GAR Module and Delineation of Microtubule Binding Determinants.
Lane, T.R., Fuchs, E., Slep, K.C.(2017) Structure 25: 1130-1138.e6
- PubMed: 28602822
- DOI: https://doi.org/10.1016/j.str.2017.05.006
- Primary Citation of Related Structures:
5VE9 - PubMed Abstract:
Spectraplakins are large molecules that cross-link F-actin and microtubules (MTs). Mutations in spectraplakins yield defective cell polarization, aberrant focal adhesion dynamics, and dystonia. We present the 2.8 Å crystal structure of the hACF7 EF1-EF2-GAR MT-binding module and delineate the GAR residues critical for MT binding. The EF1-EF2 and GAR domains are autonomous domains connected by a flexible linker. The EF1-EF2 domain is an EFβ-scaffold with two bound Ca 2+ ions that straddle an N-terminal α helix. The GAR domain has a unique α/β sandwich fold that coordinates Zn 2+ . While the EF1-EF2 domain is not sufficient for MT binding, the GAR domain is and likely enhances EF1-EF2-MT engagement. Residues in a conserved basic patch, distal to the GAR domain's Zn 2+ -binding site, mediate MT binding.
- Department of Biochemistry and Biophysics, University of North Carolina, Chapel Hill, NC 27599, USA; Molecular and Cellular Biophysics Program, University of North Carolina, Chapel Hill, NC 27599, USA.
Organizational Affiliation: