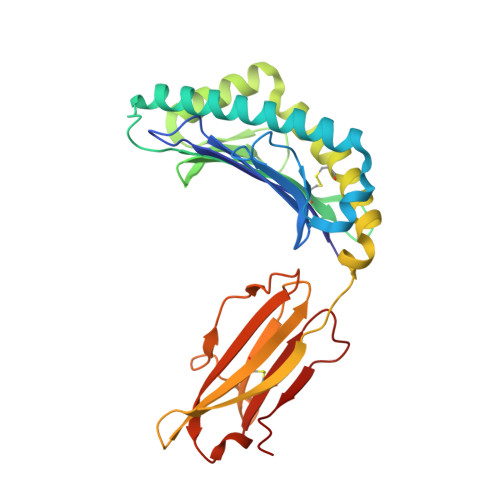
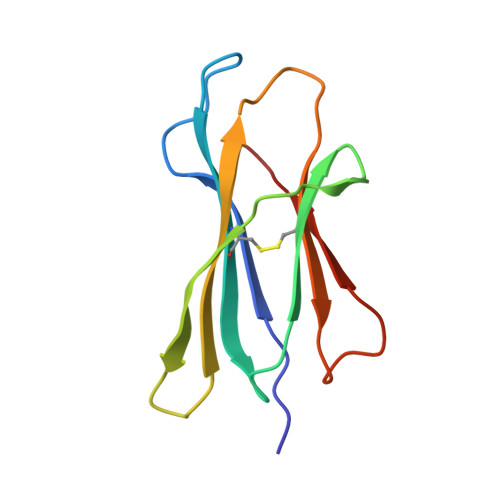
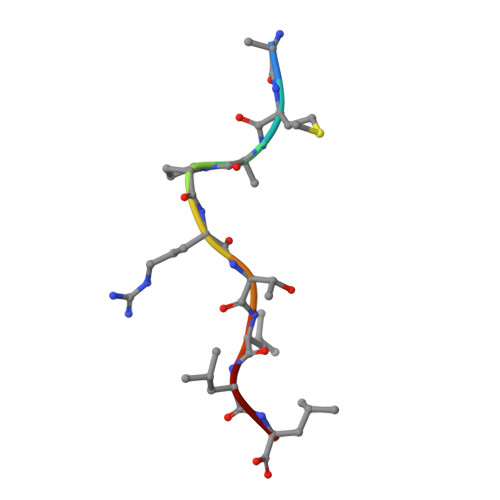
Crystal structure of Qa-1a with bound Qa-1 determinant modifier peptide.
Ying, G., Wang, J., Kumar, V., Zajonc, D.M.(2017) PLoS One 12: e0182296-e0182296
- PubMed: 28767728
- DOI: https://doi.org/10.1371/journal.pone.0182296
- Primary Citation of Related Structures:
5VCL - PubMed Abstract:
Qa-1 is a non-classical Major Histocompatibility (MHC) class I molecule that generally presents hydrophobic peptides including Qdm derived from the leader sequence of classical MHC I molecules for immune surveillance by NK cells. Qa-1 bound peptides derived from the TCR Vβ8.2 of activated T cells also activates CD8+ regulatory T cells to control autoimmunity and maintain self-tolerance. Four allotypes of Qa-1 (Qa-1a-d) are expressed that are highly conserved in sequence but have several variations that could affect peptide binding to Qa-1 or TCR recognition. Here, we determined the structure of Qa-1a with bound Qdm peptide. While the overall structure is very similar to that of Qa-1b, there are several amino acid differences around the peptide binding platform that could affect TCR recognition. Most notably, two amino acid substitutions are found in the pocket P2, which binds the anchor residue Met2 of the Qdm peptide. These residues affect both the size and shape of the binding pocket, as well as affect the charge at physiologic pH, suggesting Qa-1a and Qa-1b could present slightly distinct peptide reservoirs, which could presumably be recognized by different populations of CD8+ T cells.
- Division of Cell Biology, La Jolla Institute for Allergy and Immunology (LJI), La Jolla, California, United States of America.
Organizational Affiliation: