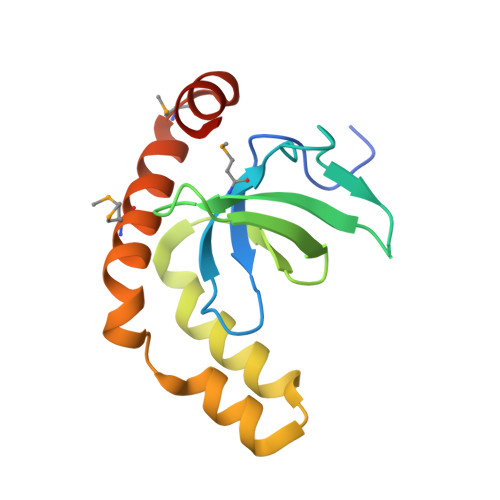
Histone and DNA binding ability studies of the NSD subfamily of PWWP domains.
Zhang, M., Yang, Y., Zhou, M., Dong, A., Yan, X., Loppnau, P., Min, J., Liu, Y.(2021) Biochem Biophys Res Commun 569: 199-206
- PubMed: 34271259
- DOI: https://doi.org/10.1016/j.bbrc.2021.07.017
- Primary Citation of Related Structures:
4RXJ, 5VC8 - PubMed Abstract:
The NSD proteins, namely NSD1, NSD2 and NSD3, are lysine methyltransferases, which catalyze mono- and di-methylation of histone H3K36. They are multi-domain proteins, including two PWWP domains (PWWP1 and PWWP2) separated by some other domains. These proteins act as potent oncoproteins and are implicated in various cancers. However the biological functions of these PWWP domains are still largely unknown. To better understand the functions of these proteins' PWWP domains, we cloned, expressed and purified all the PWWP domains of these NSD proteins to characterize their interactions with methylated histone peptides and dsDNA by quantitative binding assays and crystallographic analysis. Our studies indicate that all these PWWP domains except NSD1_PWWP1 bind to trimethylated H3K36, H3K79 peptides and dsDNA weakly. Our crystal structures uncover that the NDS3_PWWP2 and NSD2_PWWP1 domains, which hold an extremely long α-helix and α-helix bundle, respectively, need a conformation adjustment to interact with nucleosome.
- College of Pharmaceutical Sciences, Soochow University, Suzhou, Jiangsu, China.
Organizational Affiliation: