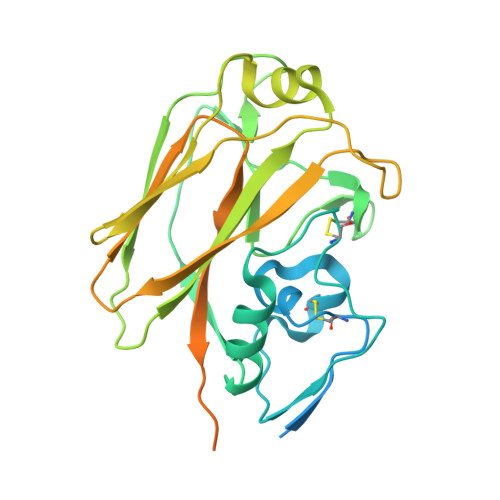
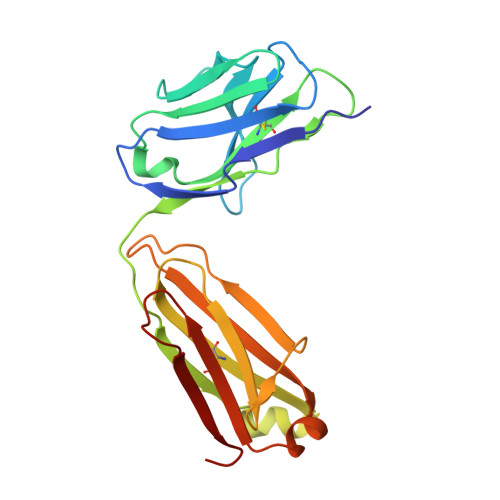
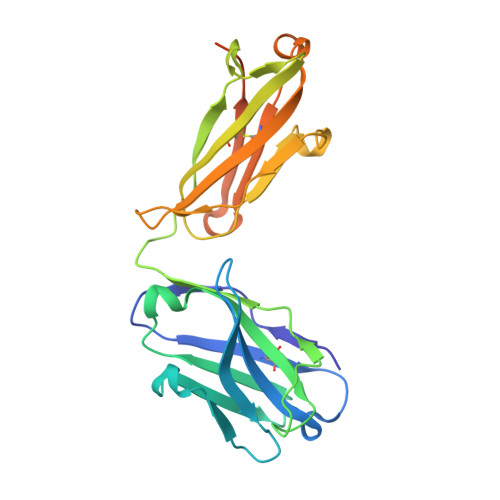
A Potent Germline-like Human Monoclonal Antibody Targets a pH-Sensitive Epitope on H7N9 Influenza Hemagglutinin.
Yu, F., Song, H., Wu, Y., Chang, S.Y., Wang, L., Li, W., Hong, B., Xia, S., Wang, C., Khurana, S., Feng, Y., Wang, Y., Sun, Z., He, B., Hou, D., Manischewitz, J., King, L.R., Song, Y., Min, J.Y., Golding, H., Ji, X., Lu, L., Jiang, S., Dimitrov, D.S., Ying, T.(2017) Cell Host Microbe 22: 471-483.e5
- PubMed: 28966056
- DOI: https://doi.org/10.1016/j.chom.2017.08.011
- Primary Citation of Related Structures:
5VAG - PubMed Abstract:
The H7N9 influenza virus causes high-mortality disease in humans but no effective therapeutics are available. Here we report a human monoclonal antibody, m826, that binds to H7 hemagglutinin (HA) and protects against H7N9 infection. m826 binds to H7N9 HA with subnanomolar affinity at acidic pH and 10-fold lower affinity at neutral pH. The high-resolution (1.9 Å) crystal structure of m826 complexed with H7N9 HA indicates that m826 binds an epitope that may be fully exposed upon pH-induced conformational changes in HA. m826 fully protects mice against lethal challenge with H7N9 virus through mechanisms likely involving antibody-dependent cell-mediated cytotoxicity. Interestingly, immunogenetic analysis indicates that m826 is a germline antibody, and m826-like sequences can be identified in H7N9-infected patients, healthy adults, and newborn babies. These m826 properties offer a template for H7N9 vaccine immunogens, a promising candidate therapeutic, and a tool for exploring mechanisms of virus infection inhibition by antibodies.
- Key Laboratory of Medical Molecular Virology of Ministries of Education and Health, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China; College of Life Sciences, Agricultural University of Hebei, Baoding, Hebei 071001, China.
Organizational Affiliation: