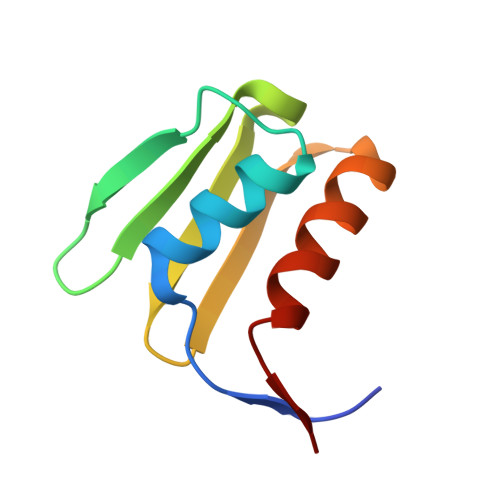
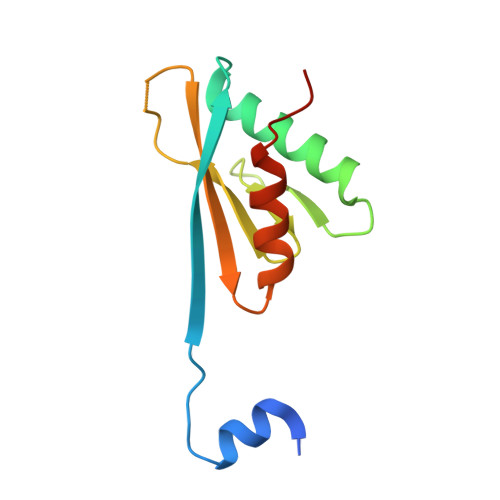
Molecular basis for the interaction between Integrator subunits IntS9 and IntS11 and its functional importance.
Wu, Y., Albrecht, T.R., Baillat, D., Wagner, E.J., Tong, L.(2017) Proc Natl Acad Sci U S A 114: 4394-4399
- PubMed: 28396433
- DOI: https://doi.org/10.1073/pnas.1616605114
- Primary Citation of Related Structures:
5V8W - PubMed Abstract:
The metazoan Integrator complex (INT) has important functions in the 3'-end processing of noncoding RNAs, including the uridine-rich small nuclear RNA (UsnRNA) and enhancer RNA (eRNA), and in the transcription of coding genes by RNA polymerase II. The INT contains at least 14 subunits, but its molecular mechanism of action is poorly understood, because currently there is little structural information about its subunits. The endonuclease activity of INT is mediated by its subunit 11 (IntS11), which belongs to the metallo-β-lactamase superfamily and is a paralog of CPSF-73, the endonuclease for pre-mRNA 3'-end processing. IntS11 forms a stable complex with Integrator complex subunit 9 (IntS9) through their C-terminal domains (CTDs). Here, we report the crystal structure of the IntS9-IntS11 CTD complex at 2.1-Å resolution and detailed, structure-based biochemical and functional studies. The complex is composed of a continuous nine-stranded β-sheet with four strands from IntS9 and five from IntS11. Highly conserved residues are located in the extensive interface between the two CTDs. Yeast two-hybrid assays and coimmunoprecipitation experiments confirm the structural observations on the complex. Functional studies demonstrate that the IntS9-IntS11 interaction is crucial for the role of INT in snRNA 3'-end processing.
- Department of Biological Sciences, Columbia University, New York, NY 10027.
Organizational Affiliation: