Structural robustness of the ribosome inferred from X-ray crystal structures of the 30S ribosomal subunit and the 70S ribosome lacking ribosomal protein uS17
Murphy, E., Connetti, J.L., Dahlberg, A.E., Gregory, S.T., Jogl, G.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Biological assembly 1 assigned by authors.
Biological assembly 2 assigned by authors.
Macromolecule Content
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L2 | C [auth 1D], CB [auth 2D] | 276 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I07 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I07 Go to UniProtKB: Q72I07 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I07 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L3 | D [auth 1E], DB [auth 2E] | 206 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I04 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I04 Go to UniProtKB: Q72I04 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I04 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L4 | E [auth 1F], EB [auth 2F] | 210 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I05 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I05 Go to UniProtKB: Q72I05 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I05 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L5 | F [auth 1G], FB [auth 2G] | 182 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I16 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I16 Go to UniProtKB: Q72I16 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I16 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L6 | G [auth 1H], GB [auth 2H] | 180 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I19 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I19 Go to UniProtKB: Q72I19 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I19 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L9 | H [auth 1I], HB [auth 2I] | 148 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72GV5 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72GV5 Go to UniProtKB: Q72GV5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72GV5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L13 | I [auth 1N], IB [auth 2N] | 140 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72IN1 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72IN1 Go to UniProtKB: Q72IN1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72IN1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L14 | J [auth 1O], JB [auth 2O] | 122 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I14 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I14 Go to UniProtKB: Q72I14 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I14 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L15 | K [auth 1P], KB [auth 2P] | 150 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I23 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I23 Go to UniProtKB: Q72I23 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I23 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L16 | L [auth 1Q], LB [auth 2Q] | 141 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I11 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I11 Go to UniProtKB: Q72I11 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I11 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L17 | M [auth 1R], MB [auth 2R] | 118 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I33 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I33 Go to UniProtKB: Q72I33 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I33 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 14 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L18 | N [auth 1S], NB [auth 2S] | 112 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I20 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I20 Go to UniProtKB: Q72I20 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I20 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 15 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L19 | O [auth 1T], OB [auth 2T] | 146 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72JU9 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72JU9 Go to UniProtKB: Q72JU9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72JU9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 16 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L20 | P [auth 1U], PB [auth 2U] | 118 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72L76 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72L76 Go to UniProtKB: Q72L76 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72L76 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 17 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L21 | Q [auth 1V], QB [auth 2V] | 101 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72HR2 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72HR2 Go to UniProtKB: Q72HR2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72HR2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 18 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L22 | R [auth 1W], RB [auth 2W] | 113 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I09 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I09 Go to UniProtKB: Q72I09 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I09 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 19 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L23 | S [auth 1X], SB [auth 2X] | 96 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I06 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I06 Go to UniProtKB: Q72I06 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I06 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 20 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L24 | T [auth 1Y], TB [auth 2Y] | 110 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I15 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I15 Go to UniProtKB: Q72I15 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I15 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 21 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L25 | U [auth 1Z], UB [auth 2Z] | 206 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72IA7 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72IA7 Go to UniProtKB: Q72IA7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72IA7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 22 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L27 | V [auth 10], VB [auth 20] | 85 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72HR3 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72HR3 Go to UniProtKB: Q72HR3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72HR3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 23 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L28 | W [auth 11], WB [auth 21] | 98 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72G84 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72G84 Go to UniProtKB: Q72G84 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72G84 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 24 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L29 | X [auth 12], XB [auth 22] | 72 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I12 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I12 Go to UniProtKB: Q72I12 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I12 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 25 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L30 | Y [auth 13], YB [auth 23] | 60 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I22 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I22 Go to UniProtKB: Q72I22 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I22 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 26 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L31 | Z [auth 14], ZB [auth 24] | 71 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72JR0 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72JR0 Go to UniProtKB: Q72JR0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72JR0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 27 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L32 | AA [auth 15], AC [auth 25] | 60 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62652 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62652 Go to UniProtKB: P62652 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62652 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 28 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L33 | BA [auth 16], BC [auth 26] | 54 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72GW3 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72GW3 Go to UniProtKB: Q72GW3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72GW3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 29 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L34 | CA [auth 17], CC [auth 27] | 49 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P80340 (Thermus thermophilus (strain ATCC 27634 / DSM 579 / HB8)) Explore P80340 Go to UniProtKB: P80340 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P80340 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 30 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L35 | DA [auth 18], DC [auth 28] | 65 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72L77 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72L77 Go to UniProtKB: Q72L77 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72L77 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 31 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L36 | EA [auth 19], EC [auth 29] | 37 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for Q72I28 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore Q72I28 Go to UniProtKB: Q72I28 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q72I28 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 33 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S2 | GA [auth 1b], GC [auth 2b] | 256 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62662 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62662 Go to UniProtKB: P62662 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62662 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 34 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S3 | HA [auth 1c], HC [auth 2c] | 239 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62663 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62663 Go to UniProtKB: P62663 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62663 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 35 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S4 | IA [auth 1d], IC [auth 2d] | 209 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62664 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62664 Go to UniProtKB: P62664 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62664 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 36 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S5 | JA [auth 1e], JC [auth 2e] | 162 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62665 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62665 Go to UniProtKB: P62665 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62665 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 37 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S6 | KA [auth 1f], KC [auth 2f] | 101 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62666 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62666 Go to UniProtKB: P62666 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62666 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 38 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S7 | LA [auth 1g], LC [auth 2g] | 156 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62667 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62667 Go to UniProtKB: P62667 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62667 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 39 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S8 | MA [auth 1h], MC [auth 2h] | 138 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62668 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62668 Go to UniProtKB: P62668 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62668 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 40 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S9 | NA [auth 1i], NC [auth 2i] | 128 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62669 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62669 Go to UniProtKB: P62669 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62669 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 41 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S10 | OA [auth 1j], OC [auth 2j] | 105 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62653 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62653 Go to UniProtKB: P62653 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62653 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 42 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S11 | PA [auth 1k], PC [auth 2k] | 129 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62654 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62654 Go to UniProtKB: P62654 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62654 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 43 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S12 | QA [auth 1l], QC [auth 2l] | 135 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for H7GHW0 (Thermus parvatiensis) Explore H7GHW0 Go to UniProtKB: H7GHW0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | H7GHW0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 44 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S13 | RA [auth 1m], RC [auth 2m] | 126 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62655 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62655 Go to UniProtKB: P62655 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62655 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 45 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S14 type Z | SA [auth 1n], SC [auth 2n] | 61 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62656 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62656 Go to UniProtKB: P62656 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62656 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 46 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S15 | TA [auth 1o], TC [auth 2o] | 89 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62657 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62657 Go to UniProtKB: P62657 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62657 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 47 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S16 | UA [auth 1p], UC [auth 2p] | 88 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62238 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62238 Go to UniProtKB: P62238 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62238 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 48 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S18 | VA [auth 1r], VC [auth 2r] | 88 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62659 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62659 Go to UniProtKB: P62659 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62659 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 49 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S19 | WA [auth 1s], WC [auth 2s] | 93 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62660 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62660 Go to UniProtKB: P62660 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62660 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 50 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S20 | XA [auth 1t], XC [auth 2t] | 106 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62661 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62661 Go to UniProtKB: P62661 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62661 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 51 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein Thx | YA [auth 1u], YC [auth 2u] | 27 | Thermus thermophilus HB27 | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62613 (Thermus thermophilus (strain ATCC BAA-163 / DSM 7039 / HB27)) Explore P62613 Go to UniProtKB: P62613 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62613 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 52 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribosome-associated inhibitor A | ZA [auth 1y], ZC [auth 2y] | 113 | Escherichia coli K-12 | Mutation(s): 0 Gene Names: raiA, yfiA, b2597, JW2578 |  |
UniProt | |||||
Find proteins for P0AD49 (Escherichia coli (strain K12)) Explore P0AD49 Go to UniProtKB: P0AD49 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AD49 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 23S Ribosomal RNA | A [auth 1A], AB [auth 2A] | 2,894 | Thermus thermophilus HB27 |  | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5S Ribosomal RNA | B [auth 1B], BB [auth 2B] | 120 | Thermus thermophilus HB27 |  | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 32 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 16S Ribosomal RNA | FA [auth 1a], FC [auth 2a] | 1,522 | Thermus thermophilus HB27 | 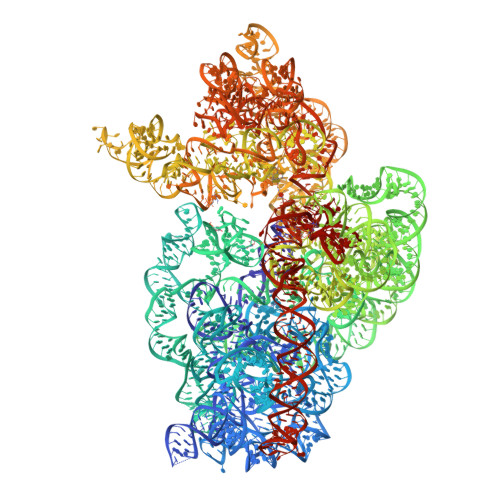 | |
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| 0TD Query on 0TD | QA [auth 1l], QC [auth 2l] | L-PEPTIDE LINKING | C5 H9 N O4 S |  | ASP |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 210.805 | α = 90 |
| b = 450.826 | β = 90 |
| c = 621.306 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Aimless | data scaling |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | GM094157 |
RCSB PDB Core Operations are funded by the U.S. National Science Foundation (DBI-2321666), the US Department of Energy (DE-SC0019749), and the National Cancer Institute, National Institute of Allergy and Infectious Diseases, and National Institute of General Medical Sciences of the National Institutes of Health under grant R01GM157729. RCSB PDB uses resources of the National Energy Research Scientific Computing Center (NERSC), a Department of Energy User Facility.





