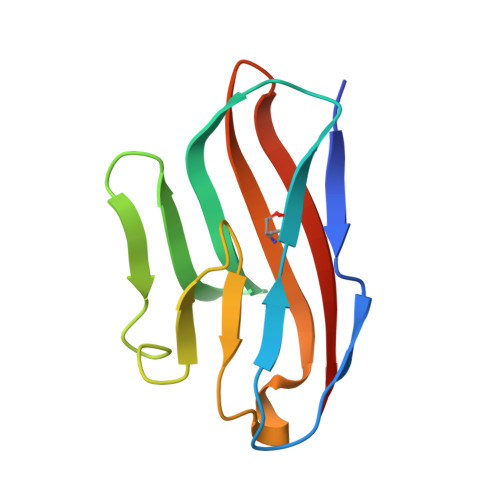
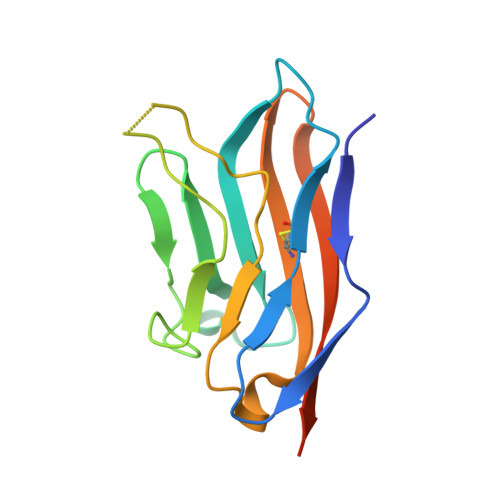
Recognition of nectin-2 by the natural killer cell receptor T cell immunoglobulin and ITIM domain (TIGIT).
Deuss, F.A., Gully, B.S., Rossjohn, J., Berry, R.(2017) J Biological Chem 292: 11413-11422
- PubMed: 28515320
- DOI: https://doi.org/10.1074/jbc.M117.786483
- Primary Citation of Related Structures:
5V52 - PubMed Abstract:
T cell immunoglobulin and ITIM domain (TIGIT) is an inhibitory receptor expressed on the surface of natural killer (NK) cells. TIGIT recognizes nectin and nectin-like adhesion molecules and thus plays a critical role in the innate immune response to malignant transformation. Although the TIGIT nectin-like protein-5 (necl-5) interaction is well understood, how TIGIT engages nectin-2, a receptor that is broadly over-expressed in breast and ovarian cancer, remains unknown. Here, we show that TIGIT bound to the immunoglobulin domain of nectin-2 that is most distal from the membrane with an affinity of 6 μm, which was moderately lower than the affinity observed for the TIGIT/necl-5 interaction (3.2 μm). The TIGIT/nectin-2 binding disrupted pre-assembled nectin-2 oligomers, suggesting that receptor-ligand and ligand-ligand associations are mutually exclusive events. Indeed, the crystal structure of TIGIT bound to the first immunoglobulin domain of nectin-2 indicated that the receptor and ligand dock using the same molecular surface and a conserved "lock and key" binding motifs previously observed to mediate nectin/nectin homotypic interactions as well as TIGIT/necl-5 recognition. Using a mutagenesis approach, we dissected the energetic basis for the TIGIT/nectin-2 interaction and revealed that an "aromatic key" of nectin-2 is critical for this interaction, whereas variations in the lock were tolerated. Moreover, we found that the C-C' loop of the ligand dictates the TIGIT binding hierarchy. Altogether, these findings broaden our understanding of nectin/nectin receptor interactions and have implications for better understanding the molecular basis for autoimmune disease and cancer.
- From the Infection and Immunity Program, Biomedicine Discovery Institute and.
Organizational Affiliation: