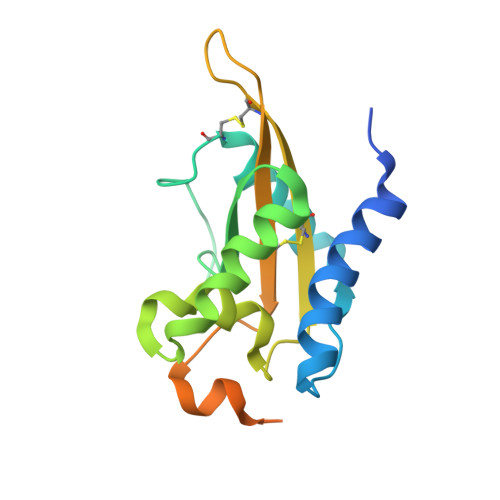
Crystal Structure of MpPR-1i, a SCP/TAPS protein from Moniliophthora perniciosa, the fungus that causes Witches' Broom Disease of Cacao.
Baroni, R.M., Luo, Z., Darwiche, R., Hudspeth, E.M., Schneiter, R., Pereira, G.A.G., Mondego, J.M.C., Asojo, O.A.(2017) Sci Rep 7: 7818-7818
- PubMed: 28798297
- DOI: https://doi.org/10.1038/s41598-017-07887-1
- Primary Citation of Related Structures:
5V50, 5V51 - PubMed Abstract:
The pathogenic fungi Moniliophthora perniciosa causes Witches' Broom Disease (WBD) of cacao. The structure of MpPR-1i, a protein expressed by M. perniciosa when it infects cacao, are presented. This is the first reported de novo structure determined by single-wavelength anomalous dispersion phasing upon soaking with selenourea. Each monomer has flexible loop regions linking the core alpha-beta-alpha sandwich topology that comprise ~50% of the structure, making it difficult to generate an accurate homology model of the protein. MpPR-1i is monomeric in solution but is packed as a high ~70% solvent content, crystallographic heptamer. The greatest conformational flexibility between monomers is found in loops exposed to the solvent channel that connect the two longest strands. MpPR-1i lacks the conserved CAP tetrad and is incapable of binding divalent cations. MpPR-1i has the ability to bind lipids, which may have roles in its infection of cacao. These lipids likely bind in the palmitate binding cavity as observed in tablysin-15, since MpPR-1i binds palmitate with comparable affinity as tablysin-15. Further studies are required to clarify the possible roles and underlying mechanisms of neutral lipid binding, as well as their effects on the pathogenesis of M. perniciosa so as to develop new interventions for WBD.
- Genomics and Expression Laboratory (LGE), Institute of Biology, CP 6109, 13083-862 UNICAMP, Campinas, Brazil.
Organizational Affiliation: