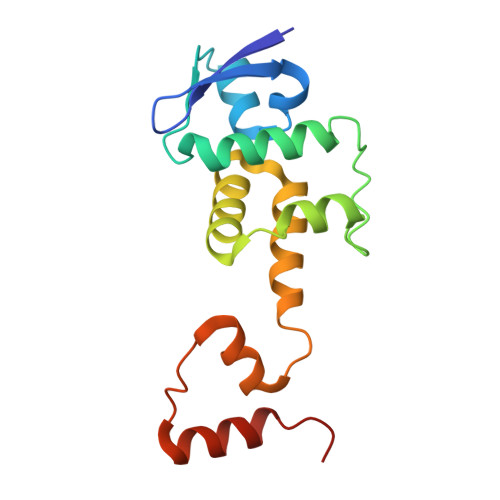
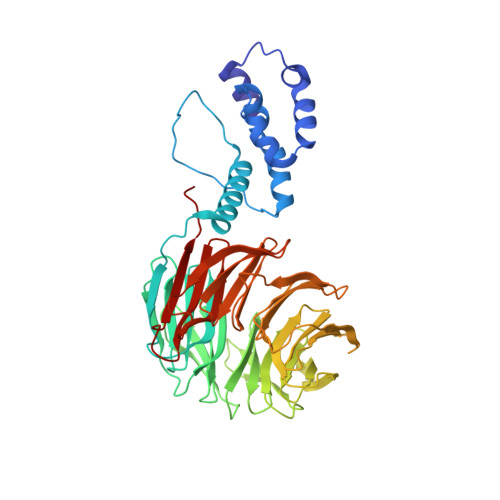
FBXW7 regulates DISC1 stability via the ubiquitin-proteosome system.
Yalla, K., Elliott, C., Day, J.P., Findlay, J., Barratt, S., Hughes, Z.A., Wilson, L., Whiteley, E., Popiolek, M., Li, Y., Dunlop, J., Killick, R., Adams, D.R., Brandon, N.J., Houslay, M.D., Hao, B., Baillie, G.S.(2018) Mol Psychiatry 23: 1278-1286
- PubMed: 28727686
- DOI: https://doi.org/10.1038/mp.2017.138
- Primary Citation of Related Structures:
5V4B - PubMed Abstract:
Disrupted in schizophrenia 1 (DISC1) is a multi-functional scaffolding protein that has been associated with neuropsychiatric disease. The role of DISC1 is to assemble protein complexes that promote neural development and signaling, hence tight control of the concentration of cellular DISC1 in neurons is vital to brain function. Using structural and biochemical techniques, we show for we believe the first time that not only is DISC1 turnover elicited by the ubiquitin proteasome system (UPS) but that it is orchestrated by the F-Box protein, FBXW7. We present the structure of FBXW7 bound to the DISC1 phosphodegron motif and exploit this information to prove that disruption of the FBXW7-DISC1 complex results in a stabilization of DISC1. This action can counteract DISC1 deficiencies observed in neural progenitor cells derived from induced pluripotent stem cells from schizophrenia patients with a DISC1 frameshift mutation. Thus manipulation of DISC1 levels via the UPS may provide a novel method to explore DISC1 function.
- College of Veterinary Medical and Life Sciences, Institute of Cardiovascular and Medical Sciences, University of Glasgow, Glasgow, UK.
Organizational Affiliation: