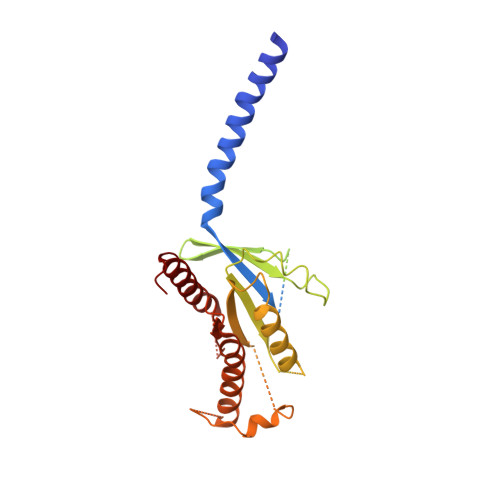
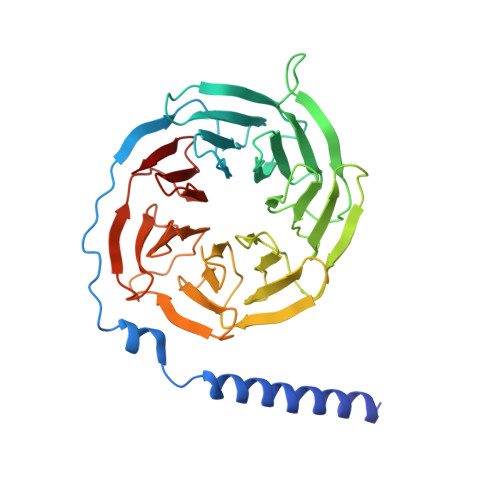
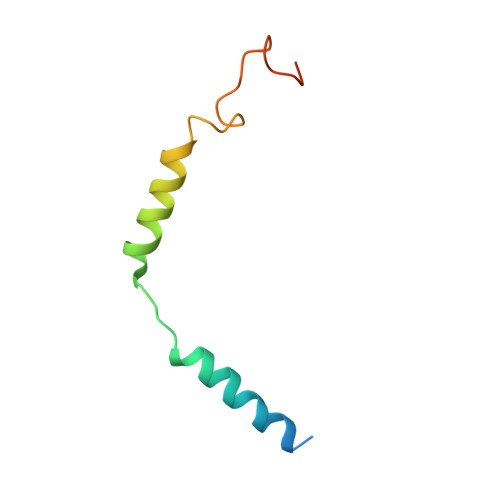
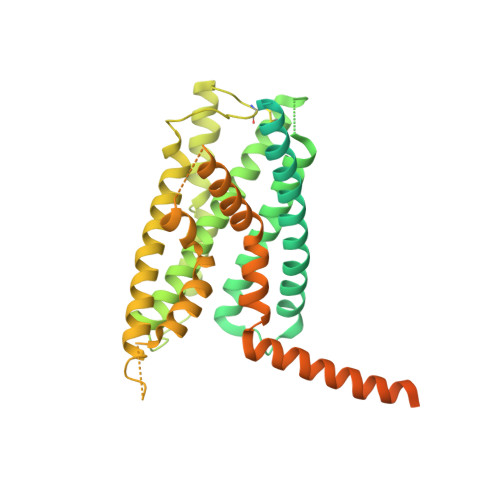
Phase-plate cryo-EM structure of a class B GPCR-G-protein complex.
Liang, Y.L., Khoshouei, M., Radjainia, M., Zhang, Y., Glukhova, A., Tarrasch, J., Thal, D.M., Furness, S.G.B., Christopoulos, G., Coudrat, T., Danev, R., Baumeister, W., Miller, L.J., Christopoulos, A., Kobilka, B.K., Wootten, D., Skiniotis, G., Sexton, P.M.(2017) Nature 546: 118-123
- PubMed: 28437792
- DOI: https://doi.org/10.1038/nature22327
- Primary Citation of Related Structures:
5UZ7 - PubMed Abstract:
Class B G-protein-coupled receptors are major targets for the treatment of chronic diseases, such as osteoporosis, diabetes and obesity. Here we report the structure of a full-length class B receptor, the calcitonin receptor, in complex with peptide ligand and heterotrimeric Gα s βγ protein determined by Volta phase-plate single-particle cryo-electron microscopy. The peptide agonist engages the receptor by binding to an extended hydrophobic pocket facilitated by the large outward movement of the extracellular ends of transmembrane helices 6 and 7. This conformation is accompanied by a 60° kink in helix 6 and a large outward movement of the intracellular end of this helix, opening the bundle to accommodate interactions with the α5-helix of Gα s . Also observed is an extended intracellular helix 8 that contributes to both receptor stability and functional G-protein coupling via an interaction with the Gβ subunit. This structure provides a new framework for understanding G-protein-coupled receptor function.
- Drug Discovery Biology and Department of Pharmacology, Monash Institute of Pharmaceutical Sciences, Monash University, Parkville, 3052 Victoria, Australia.
Organizational Affiliation: