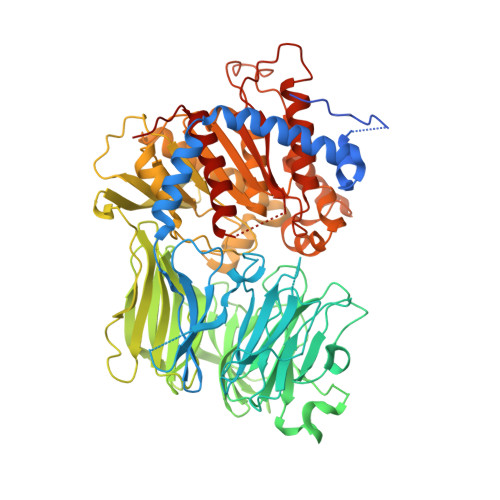
Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants.
Chekan, J.R., Estrada, P., Covello, P.S., Nair, S.K.(2017) Proc Natl Acad Sci U S A 114: 6551-6556
- PubMed: 28584123
- DOI: https://doi.org/10.1073/pnas.1620499114
- Primary Citation of Related Structures:
5UW3, 5UW5, 5UW6, 5UW7, 5UZW - PubMed Abstract:
Enzymes that can catalyze the macrocyclization of linear peptide substrates have long been sought for the production of libraries of structurally diverse scaffolds via combinatorial gene assembly as well as to afford rapid in vivo screening methods. Orbitides are plant ribosomally synthesized and posttranslationally modified peptides (RiPPs) of various sizes and topologies, several of which are shown to be biologically active. The diversity in size and sequence of orbitides suggests that the corresponding macrocyclases may be ideal catalysts for production of cyclic peptides. Here we present the biochemical characterization and crystal structures of the plant enzyme PCY1 involved in orbitide macrocyclization. These studies demonstrate how the PCY1 S9A protease fold has been adapted for transamidation, rather than hydrolysis, of acyl-enzyme intermediates to yield cyclic products. Notably, PCY1 uses an unusual strategy in which the cleaved C-terminal follower peptide from the substrate stabilizes the enzyme in a productive conformation to facilitate macrocyclization of the N-terminal fragment. The broad substrate tolerance of PCY1 can be exploited as a biotechnological tool to generate structurally diverse arrays of macrocycles, including those with nonproteinogenic elements.
- Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, IL 61801.
Organizational Affiliation: