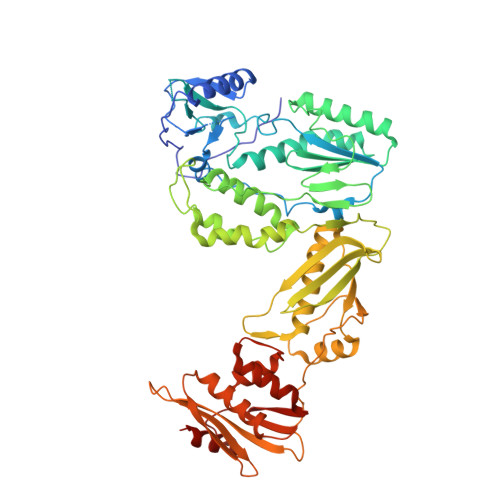
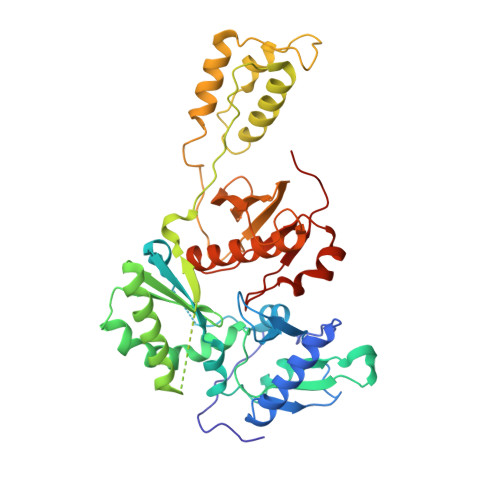
A 2-Hydroxyisoquinoline-1,3-Dione Active-Site RNase H Inhibitor Binds in Multiple Modes to HIV-1 Reverse Transcriptase.
Kirby, K.A., Myshakina, N.A., Christen, M.T., Chen, Y.L., Schmidt, H.A., Huber, A.D., Xi, Z., Kim, S., Rao, R.K., Kramer, S.T., Yang, Q., Singh, K., Parniak, M.A., Wang, Z., Ishima, R., Sarafianos, S.G.(2017) Antimicrob Agents Chemother 61
- PubMed: 28760905
- DOI: https://doi.org/10.1128/AAC.01351-17
- Primary Citation of Related Structures:
5UV5 - PubMed Abstract:
The RNase H (RNH) function of HIV-1 reverse transcriptase (RT) plays an essential part in the viral life cycle. We report the characterization of YLC2-155, a 2-hydroxyisoquinoline-1,3-dione (HID)-based active-site RNH inhibitor. YLC2-155 inhibits both polymerase (50% inhibitory concentration [IC 50 ] = 2.6 μM) and RNH functions (IC 50 = 0.65 μM) of RT but is more effective against RNH. X-ray crystallography, nuclear magnetic resonance (NMR) analysis, and molecular modeling were used to show that YLC2-155 binds at the RNH-active site in multiple conformations.
- Christopher S. Bond Life Sciences Center, University of Missouri, Columbia, Missouri, USA kirbyk@missouri.edu sarafianoss@missouri.edu.
Organizational Affiliation: