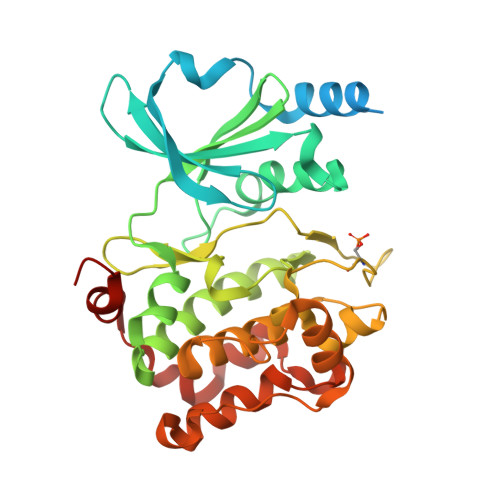
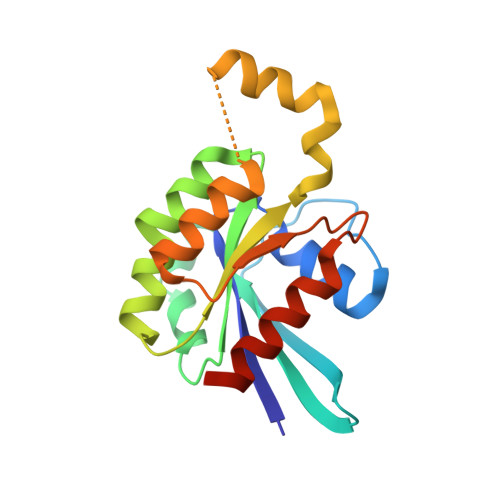
CDC42 binds PAK4 via an extended GTPase-effector interface.
Ha, B.H., Boggon, T.J.(2018) Proc Natl Acad Sci U S A 115: 531-536
- PubMed: 29295922
- DOI: https://doi.org/10.1073/pnas.1717437115
- Primary Citation of Related Structures:
5UPK, 5UPL - PubMed Abstract:
The p21-activated kinase (PAK) group of serine/threonine kinases are downstream effectors of RHO GTPases and play important roles in regulation of the actin cytoskeleton, cell growth, survival, polarity, and development. Here we probe the interaction of the type II PAK, PAK4, with RHO GTPases. Using solution scattering we find that the full-length PAK4 heterodimer with CDC42 adopts primarily a compact organization. X-ray crystallography reveals the molecular nature of the interaction between PAK4 and CDC42 and shows that in addition to the canonical PAK4 CDC42/RAC interactive binding (CRIB) domain binding to CDC42 there are unexpected contacts involving the PAK4 kinase C-lobe, CDC42, and the PAK4 polybasic region. These additional interactions modulate kinase activity and increase the binding affinity of CDC42 for full-length PAK4 compared with the CRIB domain alone. We therefore show that the interaction of CDC42 with PAK4 can influence kinase activity in a previously unappreciated manner.
- Department of Pharmacology, Yale University School of Medicine, New Haven, CT 06520.
Organizational Affiliation: