Crystal Structure Analysis of Elbow-Engineered-Fab-Bound Human Insulin Degrading Enzyme (IDE)
Liang, W.G., Bailey, L., Kossiakoff, T., Tang, W.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Insulin-degrading enzyme | 990 | Homo sapiens | Mutation(s): 13 Gene Names: IDE EC: 3.4.24.56 | 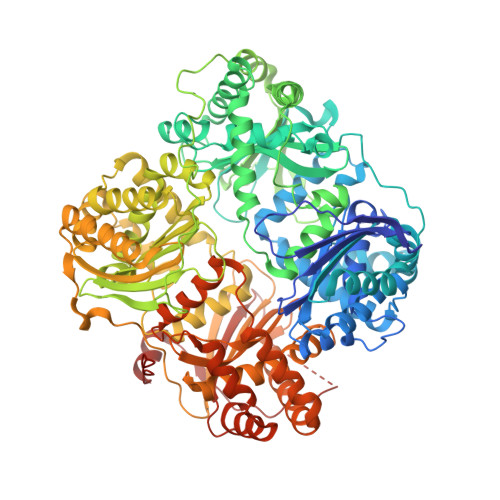 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P14735 (Homo sapiens) Explore P14735 Go to UniProtKB: P14735 | |||||
PHAROS: P14735 GTEx: ENSG00000119912 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P14735 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| FAB Heavy chain with engineered elbow | F [auth H], H [auth M], J [auth P], L [auth S], N [auth V] | 229 | Homo sapiens | Mutation(s): 0 | 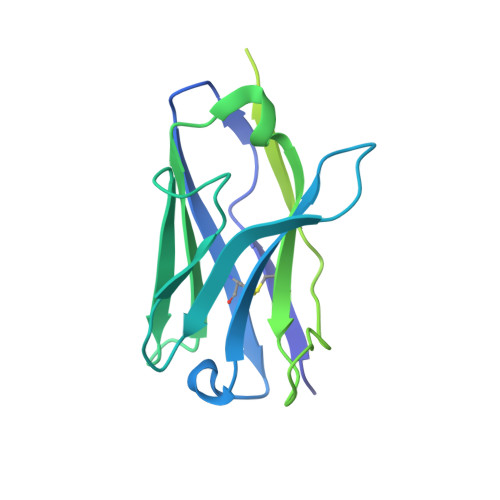 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| FAB light chain | G [auth L], I [auth N], K [auth Q], M [auth T], O [auth W] | 215 | Homo sapiens | Mutation(s): 0 | 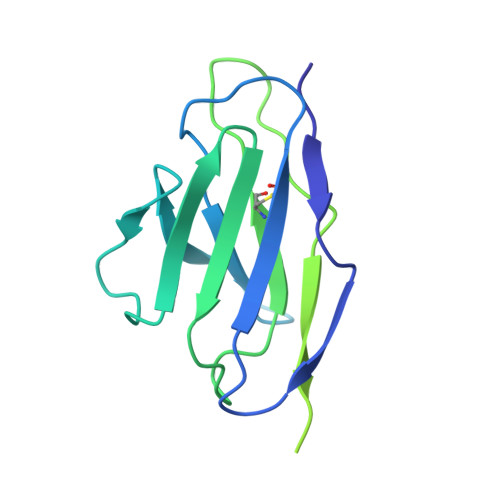 |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 131.321 | α = 90 |
| b = 242.048 | β = 90 |
| c = 310.757 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |