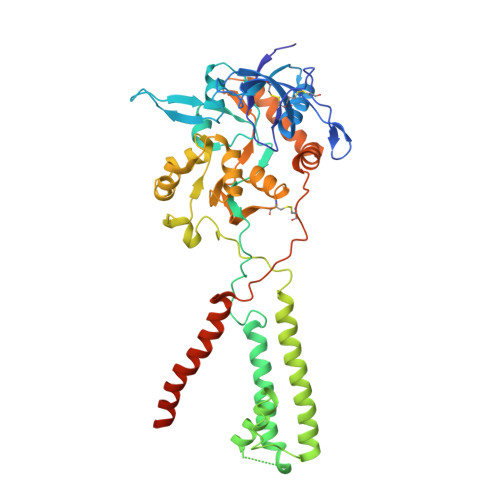
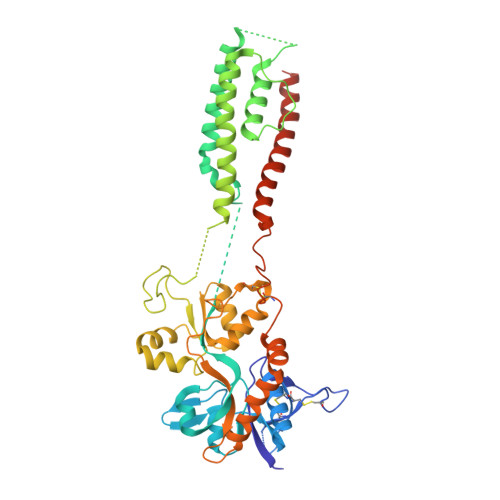
Mechanism of NMDA receptor channel block by MK-801 and memantine.
Song, X., Jensen, M.O., Jogini, V., Stein, R.A., Lee, C.H., Mchaourab, H.S., Shaw, D.E., Gouaux, E.(2018) Nature 556: 515-519
- PubMed: 29670280
- DOI: https://doi.org/10.1038/s41586-018-0039-9
- Primary Citation of Related Structures:
5UN1 - PubMed Abstract:
The NMDA (N-methyl-D-aspartate) receptor transduces the binding of glutamate and glycine, coupling it to the opening of a calcium-permeable ion channel 1 . Owing to the lack of high-resolution structural studies of the NMDA receptor, the mechanism by which ion-channel blockers occlude ion permeation is not well understood. Here we show that removal of the amino-terminal domains from the GluN1-GluN2B NMDA receptor yields a functional receptor and crystals with good diffraction properties, allowing us to map the binding site of the NMDA receptor blocker, MK-801. This crystal structure, together with long-timescale molecular dynamics simulations, shows how MK-801 and memantine (a drug approved for the treatment of Alzheimer's disease) bind within the vestibule of the ion channel, promote closure of the ion channel gate and lodge between the M3-helix-bundle crossing and the M2-pore loops, physically blocking ion permeation.
- Vollum Institute, Oregon Health & Science University, Portland, OR, USA.
Organizational Affiliation: