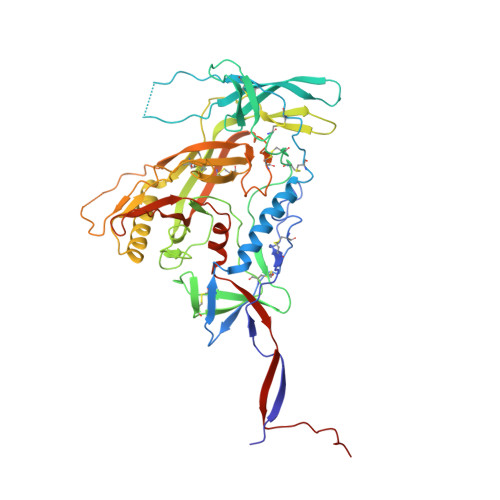
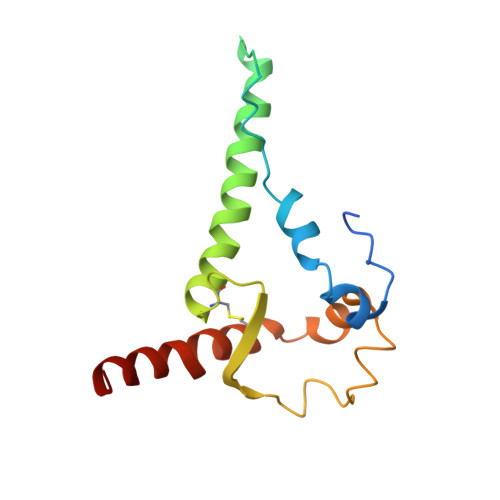
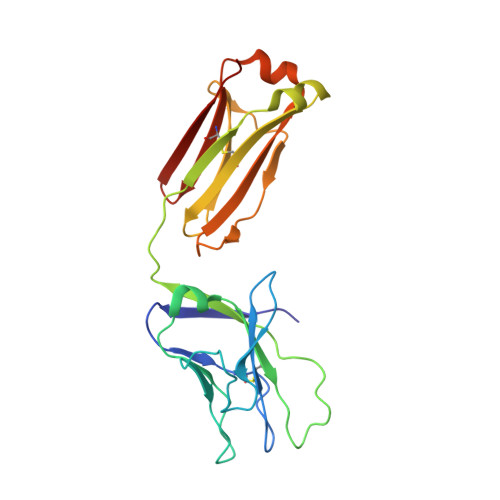
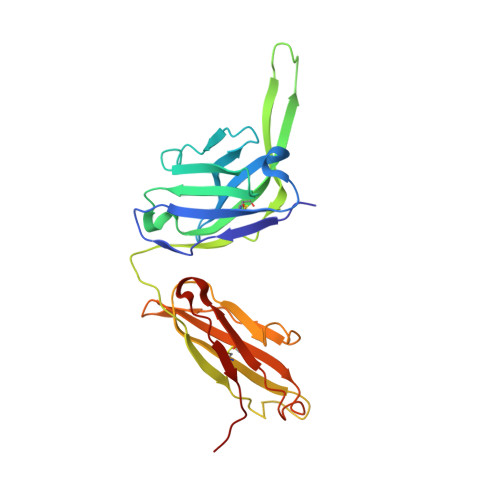
Glycine Substitution at Helix-to-Coil Transitions Facilitates the Structural Determination of a Stabilized Subtype C HIV Envelope Glycoprotein.
Guenaga, J., Garces, F., de Val, N., Stanfield, R.L., Dubrovskaya, V., Higgins, B., Carrette, B., Ward, A.B., Wilson, I.A., Wyatt, R.T.(2017) Immunity 46: 792-803.e3
- PubMed: 28514686
- DOI: https://doi.org/10.1016/j.immuni.2017.04.014
- Primary Citation of Related Structures:
5UM8 - PubMed Abstract:
Advances in HIV-1 envelope glycoprotein (Env) design generate native-like trimers and high-resolution clade A, B, and G structures and elicit neutralizing antibodies. However, a high-resolution clade C structure is critical, as this subtype accounts for the majority of HIV infections worldwide, but well-ordered clade C Env trimers are more challenging to produce due to their instability. Based on targeted glycine substitutions in the Env fusion machinery, we defined a general approach that disfavors helical transitions leading to post-fusion conformations, thereby favoring the pre-fusion state. We generated a stabilized, soluble clade C Env (16055 NFL) and determined its crystal structure at 3.9 Å. Its overall conformation is similar to SOSIP.664 and native Env trimers but includes a covalent linker between gp120 and gp41, an engineered 201-433 disulfide bond, and density corresponding to 22 N-glycans. Env-structure-guided design strategies resulted in multiple homogeneous cross-clade immunogens with the potential to advance HIV vaccine development.
- IAVI Neutralizing Antibody Center at The Scripps Research Institute, La Jolla, CA 92037, USA.
Organizational Affiliation: