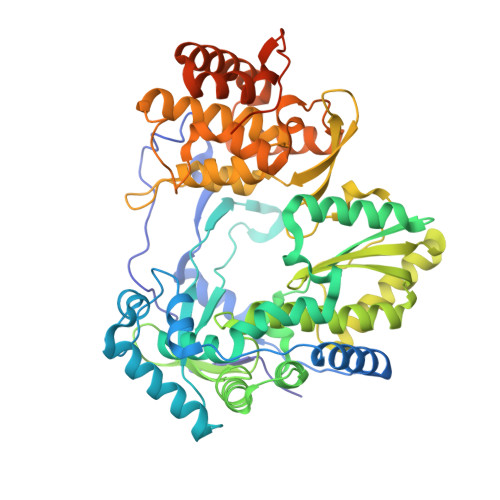
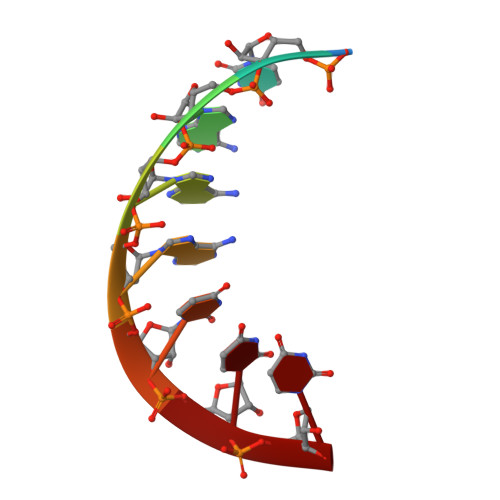
Discovery of a 2'-fluoro-2'-C-methyl C-nucleotide HCV polymerase inhibitor and a phosphoramidate prodrug with favorable properties.
Kirschberg, T.A., Metobo, S., Clarke, M.O., Aktoudianakis, V., Babusis, D., Barauskas, O., Birkus, G., Butler, T., Byun, D., Chin, G., Doerffler, E., Edwards, T.E., Fenaux, M., Lee, R., Lew, W., Mish, M.R., Murakami, E., Park, Y., Squires, N.H., Tirunagari, N., Wang, T., Whitcomb, M., Xu, J., Yang, H., Ye, H., Zhang, L., Appleby, T.C., Feng, J.Y., Ray, A.S., Cho, A., Kim, C.U.(2017) Bioorg Med Chem Lett 27: 1840-1847
- PubMed: 28274633
- DOI: https://doi.org/10.1016/j.bmcl.2017.02.037
- Primary Citation of Related Structures:
5UJ2 - PubMed Abstract:
A series of 2'-fluorinated C-nucleosides were prepared and tested for anti-HCV activity. Among them, the triphosphate of 2'-fluoro-2'-C-methyl adenosine C-nucleoside (15) was a potent and selective inhibitor of the NS5B polymerase and maintained activity against the S282T resistance mutant. A number of phosphoramidate prodrugs were then prepared and evaluated leading to the identification of the 1-aminocyclobutane-1-carboxylic acid isopropyl ester variant (53) with favorable pharmacokinetic properties including efficient liver delivery in animals.
- Gilead Sciences, 333 Lakeside Drive, Foster City, CA 94404, USA. Electronic address: tkirschberg@gilead.com.
Organizational Affiliation: